GgaEX0009303 @ galGal4
Exon Skipping
Gene
ENSGALG00000006531 | TRA2B
Description
transformer-2 protein homolog beta [Source:RefSeq peptide;Acc:NP_990009]
Coordinates
chr9:3895926-3901183:+
Coord C1 exon
chr9:3895926-3896043
Coord A exon
chr9:3900517-3900794
Coord C2 exon
chr9:3901050-3901183
Length
278 bp
Sequences
Splice sites
3' ss Seq
CCTGTTCTTTTTCTATTAAGGTT
3' ss Score
10.29
5' ss Seq
TAAGTAATT
5' ss Score
1.76
Exon sequences
Seq C1 exon
ACTTCCTCTGCGGAGAGCGCTGGCGCCGGGGACGGTCGGGTCGGGTCGGCGCGGAGGTGGCGGGGAGTGGCGCGGCGGGACGATGAGCGACAGCGGGGAGCAGAACTACGGCGAGCGG
Seq A exon
GTTAATGTTGAAGAAGGAAAATGCGGAAGTCGCCATTTGACAAGTTTTATAAATGAGAATTATTTGAAGCTCAGGAATAAGTGAAGCTGAAATTTGAAAAAAAAGAAAGTGAATGCATGCTAATTATCAGACCAGAAGTCCCACTTGTAGAATATTGAGCAATTTGTGTGAAGTGGGGAAGATGAAAGAAGTCAGAATTTGAAACGGAAGAATGAAGAAAAGGAATAAAAATGAAGTTAAGATAAGAAGTAATCTGGAATCTGAAAGCACTACGCTAA
Seq C2 exon
GAATCCCGTTCTGCTTCCAGAAGCGGAAGTGCTCATGGGTCTGGCAAGTCGGGGAGGCACACCCCTGCAAGATCTCGATCTAAGGAGGATTCAAGGCGTTCCAGGTCAAAATCTAGATCCAGGTCTGAATCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000006531_MULTIEX1-1/2=C1-2
Average complexity
C1*
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
Show structural model
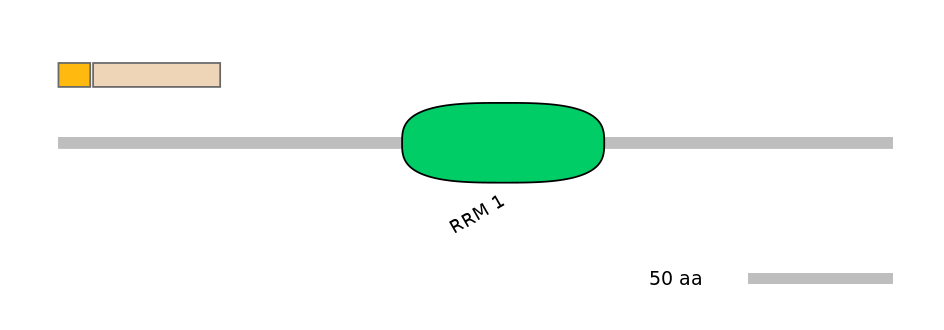
Features
Disorder rate (Iupred):
C1=1.000 A=0.185 C2=1.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Inclusion Isoform:
ENSGALT00000010555fB1021
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACTTCCTCTGCGGAGAGC
R:
CTGGATTCAGACCTGGATCTAG
Band lengths:
252-530
Functional annotations
There are 4 annotated functions for this event
PMID: 14709600
TRA2-BETA1 (canonical isoform) binds to four enhancers present in exon 2 HsaEX0066781), which activates its inclusion. Inclusion of exon 2 generates mRNAs that are not translated into proteins. HTRA2-BETA1 interacting proteins promote exon 2 skipping by sequestering it, which upregulates the HTRA2-BETA1 protein synthesis. It is proposed that the regulation of the tra2-beta pre-mRNA alternative splicing provides a robust and sensitive molecular sensor that measures the ratio between HTRA2-BETA1 and its interacting proteins.
PMID: 31911676
[CRISPR screen]. Conserved poison exon with mixed impact when depleted: HeLa Enriched at 14 days (FC=1.571, FDR=0.000), PC9 No Change at 14 days (FC=0.954, FDR=0.648), Late Xenograft Depleted (FC=0.512, FDR=0.000).
PMID: 33176162
Poison exons in serine-arginine-rich (SR) proteins, a family of 14 essential SFs, are differentially spliced during induced pluripotent stem cell (iPSC) differentiation and in tumors versus normal tissues. The study uncovers an extensive cross-regulatory network of SR proteins controlling their expression via alternative splicing coupled to nonsense-mediated decay.
PMID: 33176162
Splice-switching antisense oligonucleotides that reverse the increased skipping of TRA2B-Poison exon detected in breast tumors, alters breast cancer cell viability, proliferation, and migration.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]