GgaEX1007370 @ galGal4
Exon Skipping
Gene
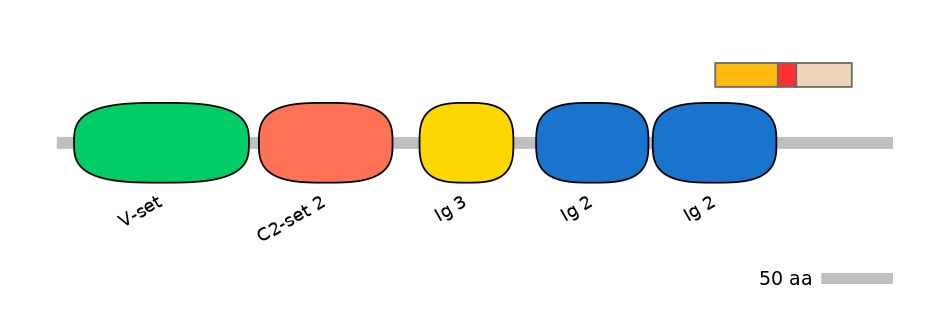
ENSGALG00000015348 | ALCAM
Description
CD166 antigen [Source:UniProtKB/Swiss-Prot;Acc:P42292]
Coordinates
chr1:85997695-85999041:+
Coord C1 exon
chr1:85997695-85997827
Coord A exon
chr1:85998566-85998604
Coord C2 exon
chr1:85998924-85999041
Length
39 bp
Sequences
Splice sites
3' ss Seq
TTTAATGTTTTTCATTTCAGTAA
3' ss Score
8.17
5' ss Seq
ATGGTATGT
5' ss Score
8.35
Exon sequences
Seq C1 exon
ACAGAGGAGACCAAATATGTTAATGGAAAATTTTCCAGTAAAATTATAATTGCTCCTGAGGAAAACGTAACATTAACATGCATTGCAGAAAATGAGCTAGAAAGGACTGTGACCTCTCTGAATGTCTCTGCTA
Seq A exon
TAAGTATTCCAGAATATGATGAACCGGAGGATAGAAATG
Seq C2 exon
ATGACAACAGCGAAAAGGTTAATGACCAGGCAAAGCTAATCGTGGGGATTGTAGTCGGTCTTCTGCTGGTTGCCCTGGTTGCAGGTGTTGTCTACTGGCTCTATGTGAAGAAATCAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000015348-'23-18,'23-17,24-18
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.786 C2=0.150
Domain overlap (PFAM):
C1:
PF138951=Ig_2=PD(48.9=95.6),PF142831=DUF4366=PU(28.3=71.1)
A:
PF142831=DUF4366=FE(11.5=100)
C2:
PF142831=DUF4366=FE(34.5=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGCTCCTGAGGAAAACGTAACA
R:
CCACGATTAGCTTTGCCTGGT
Band lengths:
128-167
Functional annotations
There are 2 annotated functions for this event
PMID: 29453336
Loss of the membrane-proximal region of ALCAM (exon 13) increased metastasis four-fold. Mechanistic studies identified a novel MMP14-dependent membrane distal cleavage site in ALCAM-Iso2, which mediated a ten-fold increase in shedding, thereby decreasing cellular cohesion.
PMID: 32249828
[CRISPR screen]. This alternative exon is implicated in cell fitness by a CRISPR-based exon deletion phenotypic screen in RPE-1 cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]