MmuEX0004576 @ mm10
Exon Skipping
Gene
ENSMUSG00000022636 | Alcam
Description
activated leukocyte cell adhesion molecule [Source:MGI Symbol;Acc:MGI:1313266]
Coordinates
chr16:52268382-52270779:-
Coord C1 exon
chr16:52270647-52270779
Coord A exon
chr16:52268790-52268828
Coord C2 exon
chr16:52268382-52268499
Length
39 bp
Sequences
Splice sites
3' ss Seq
ATTAACATTTTTCATTTCAGTAA
3' ss Score
7.01
5' ss Seq
GTGGTAGGT
5' ss Score
7.23
Exon sequences
Seq C1 exon
ACAGAGGAGTCTCCTTATATTAATGGCAGGTATTATAGTAAAATTATCATTTCCCCTGAGGAGAATGTAACATTAACTTGCACAGCAGAAAACCAACTGGAGAGAACAGTAAACTCCCTGAATGTCTCTGCGA
Seq A exon
TAAGTATTCCAGAGCACGATGAGGCAGACGATATAAGTG
Seq C2 exon
ATGAAAACAGAGAAAAGGTGAATGACCAGGCAAAACTGATTGTGGGAATTGTCGTTGGTCTCCTCCTCGCTGCCCTCGTTGCTGGTGTCGTCTACTGGCTGTACATGAAGAAGTCCAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000022636_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.093 A=0.564 C2=0.088
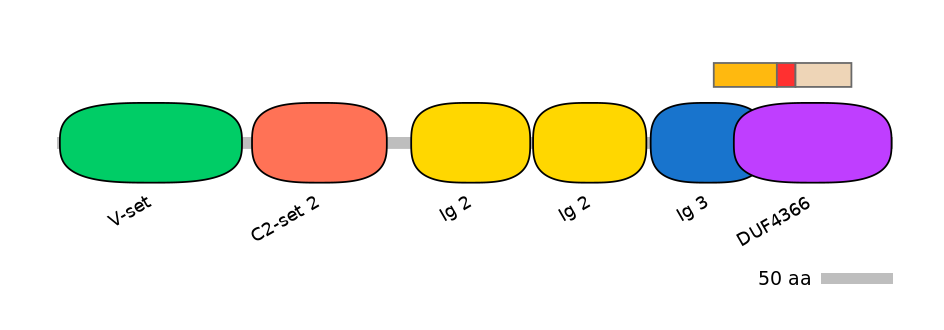
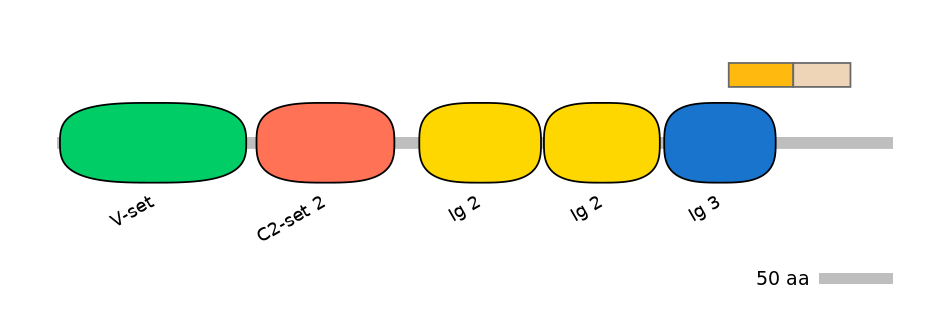
Domain overlap (PFAM):
C1:
PF139271=Ig_3=PD(44.4=80.0),PF142831=DUF4366=PU(27.0=66.7)
A:
PF142831=DUF4366=FE(11.7=100)
C2:
PF142831=DUF4366=FE(35.1=100)
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACTCCCTGAATGTCTCTGCGA
R:
GTAGACGACACCAGCAACGAG
Band lengths:
116-155
Functional annotations
There are 2 annotated functions for this event
PMID: 29453336
Loss of the membrane-proximal region of ALCAM (exon 13) increased metastasis four-fold. Mechanistic studies identified a novel MMP14-dependent membrane distal cleavage site in ALCAM-Iso2, which mediated a ten-fold increase in shedding, thereby decreasing cellular cohesion.
PMID: 32249828
[CRISPR screen]. This alternative exon is implicated in cell fitness by a CRISPR-based exon deletion phenotypic screen in RPE-1 cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types