GgaEX1066072 @ galGal4
Exon Skipping
Gene
ENSGALG00000003578 | fn1
Description
fibronectin precursor [Source:RefSeq peptide;Acc:NP_001185641]
Coordinates
chr7:4375280-4378555:-
Coord C1 exon
chr7:4378418-4378555
Coord A exon
chr7:4376687-4376845
Coord C2 exon
chr7:4375280-4375480
Length
159 bp
Sequences
Splice sites
3' ss Seq
TAATCATGTTCCTTGTGCAGATA
3' ss Score
8.84
5' ss Seq
CTGGTAAGT
5' ss Score
10.65
Exon sequences
Seq C1 exon
CCGAGCGGTGCTACGACAACACGGCCGGGACATCCTATGTGGTGGGTGAGACCTGGGAGAAGCCCTACCAGGGCTGGATGATGGTGGACTGCACCTGCCTGGGGGAGGGCAGCGGCCGCATCACCTGTACCTCCAGAA
Seq A exon
ATAGGTGCAATGACCAAGACACCAAGACTTCCTACAGAATTGGGGACACTTGGAGCAAGAAGGATAACCGGGGAAACTTGCTTCAGTGCATCTGTACTGGTAACGGCAGAGGTGAATGGAAATGCGAAAGGCACACATCCCTGCATACAACTAGCACTG
Seq C2 exon
GATCTGGCTCTCCCAGCTTCACAAATGTGCAGACTGCGTTGTACCAGCCTCAGCCTCAACAGCCCCAACCTCAGCCACATGGCCACTGTGTAACTGATAATGGAGTGGTGTATTCTTTGGGTATGCAGTGGTTGAAGACACAGGGCAGCCAGCAAATGCTTTGTACTTGTCTGGGCAATGGAGTCAGCTGCCAGGAAATAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000003578-'6-7,'6-5,8-7
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
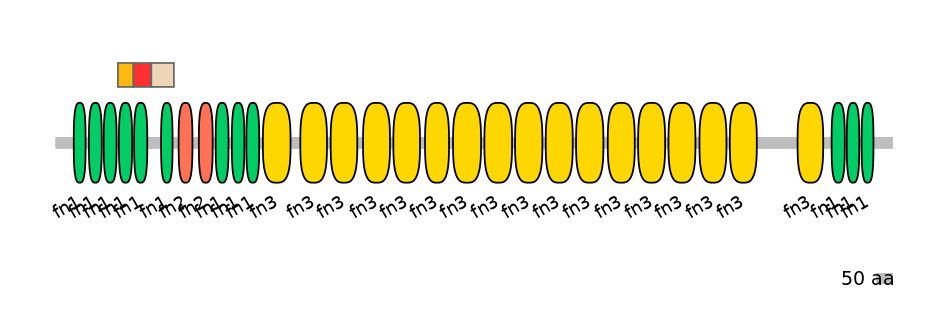
Features
Disorder rate (Iupred):
C1=0.000 A=0.093 C2=0.382
Domain overlap (PFAM):
C1:
PF0003913=fn1=WD(100=85.1)
A:
PF0003913=fn1=WD(100=74.1)
C2:
PF0003913=fn1=WD(100=51.5)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCTATGTGGTGGGTGAGACCT
R:
CCTGTGTCTTCAACCACTGCA
Band lengths:
249-408
Functional annotations
There are 6 annotated functions for this event
PMID: 1532572
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: affinity chromatography technology, coimmunoprecipitation, polyclonal antibody, western blot. ELM ID: ELMI002206; ELM sequence: NGR; Overlap: FULL
PMID: 1694173
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: affinity chromatography technology, coimmunoprecipitation, polyclonal antibody, western blot. ELM ID: ELMI002206; ELM sequence: NGR; Overlap: FULL
PMID: 17015452
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: affinity chromatography technology, coimmunoprecipitation, nuclear magnetic resonance, peptide binding to perpetrator, polyclonal antibody, western blot. ELM ID: ELMI002206; ELM sequence: NGR; Overlap: FULL
PMID: 18480047
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: nuclear magnetic resonance. ELM ID: ELMI002206; ELM sequence: NGR; Overlap: FULL
PMID: 17591922
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: nuclear magnetic resonance, sequence based prediction. ELM ID: ELMI002210; ELM sequence: NGR; Overlap: FULL
PMID: 18480047
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: nuclear magnetic resonance. ELM ID: ELMI002210; ELM sequence: NGR; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]