HsaEX0002240 @ hg19
Exon Skipping
Gene
ENSG00000186529 | AD000685.1
Description
cytochrome P450, family 4, subfamily F, polypeptide 3 [Source:HGNC Symbol;Acc:2646]
Coordinates
chr19:15752225-15757915:+
Coord C1 exon
chr19:15752225-15752423
Coord A exon
chr19:15756529-15756673
Coord C2 exon
chr19:15757862-15757915
Length
145 bp
Sequences
Splice sites
3' ss Seq
TCCTGTGTCTTTCTCTCCAGATT
3' ss Score
12.16
5' ss Seq
CAGGTAGAC
5' ss Score
6.89
Exon sequences
Seq C1 exon
GATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGGCAGCATCCCCGTGGCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACTGCTGCCGCCTCCGGTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTG
Seq A exon
ATTCACAGCTCGGAGGAAGGTCTCCTATACACACAAAGCCTGGCATGCACCTTCGGTGATATGTGCTGCTGGTGGGTGGGGCCCTGGCACGCAATCGTCCGCATCTTCCACCCCACCTACATCAAGCCTGTGCTCTTTGCTCCAG
Seq C2 exon
CTGCCATTGTACCAAAGGACAAGGTCTTCTACAGCTTCCTGAAGCCCTGGCTGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000186529_MULTIEX2-2/5=C1-3
Average complexity
ME(1-2[100=96])
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
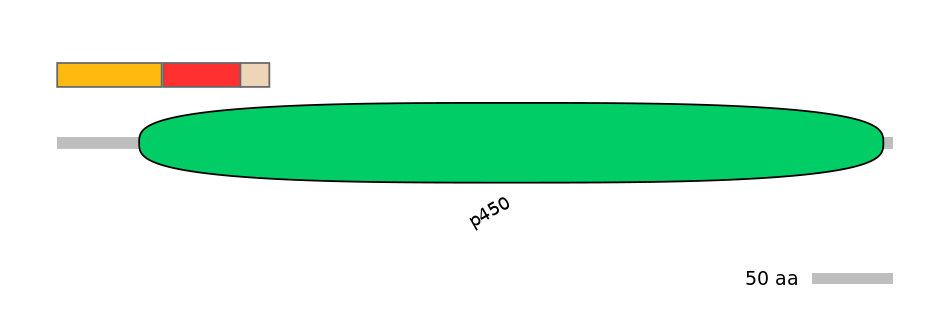
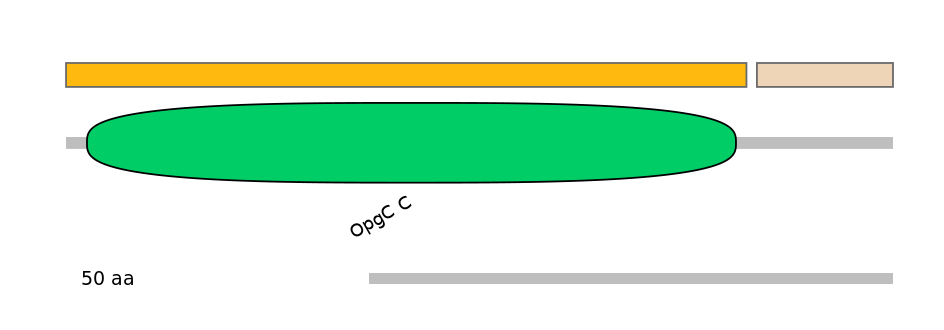
Domain overlap (PFAM):
C1:
PF0006717=p450=PU(3.0=21.2)
A:
PF0006717=p450=FE(10.3=100)
C2:
PF0006717=p450=FE(3.9=100)
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GATGCCACAGCTGAGCCTG
R:
TTCAGGAAGCTGTAGAAGACCT
Band lengths:
242-387
Functional annotations
There are 3 annotated functions for this event
PMID: 10409674
This event
Cytochrome P450 4F3 (CYP4F3) catalyzes the inactivation of leukotriene B(4) by omega-oxidation in human neutrophils. The CYP4F3 gene contains 14 exons and 13 introns. The cDNAs for CYP4F3A (the neutrophil isoform) and CYP4F3B have identical coding regions, except that they contain exons 4 (HsaEX0002240) and 3 (HsaEX0002239), respectively. Both exons code for amino acids 66-114 but share only 27% identity. When expressed in COS-7 cells, the K(m) of CYP4F3B was determined to be 26-fold higher than the K(m) of CYP4F3A using leukotriene B(4) as a substrate.
PMID: 11461919
This event
Substrate specificity. Mutually exclusive exons, each coding for 48 amino acids, generate isoforms of human CYP4F3 that differ in substrate specificity, tissue distribution, and biological function. Isoform with HsaEX0002240 inactivates LTB4 by omega-hydroxylation (Km = 0.68 microm) but has low activity for arachidonic acid (Km = 185 microm); it is the only CYP4F isoform expressed in myeloid cells in peripheral blood and bone marrow.
PMID: 15145985
This event
Among recombinant P450 enzymes, CYP4F2 and CYP4F3B (uses mutually exclusive exon HsaEX0002239) catalyzed mainly the omega-hydroxylation of C18-epoxides with an apparent Vmax of between 0.84 and 15.0 min_1, whereas the apparent Vmax displayed by CYP4F3A, the isoform found in leukocytes (uses mutually exclusive exon HsaEX0002240), ranged from 3.0 to 21.2 min_1.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)