HsaEX0004099 @ hg19
Exon Skipping
Gene
ENSG00000091879 | ANGPT2
Description
angiopoietin 2 [Source:HGNC Symbol;Acc:485]
Coordinates
chr8:6385076-6420930:-
Coord C1 exon
chr8:6420168-6420930
Coord A exon
chr8:6389853-6390008
Coord C2 exon
chr8:6385076-6385197
Length
156 bp
Sequences
Splice sites
3' ss Seq
CTAACACTGTTCTTTTCTAGCTT
3' ss Score
8.62
5' ss Seq
CAAGTAAGT
5' ss Score
10.08
Exon sequences
Seq C1 exon
CACTGCAATCTGACAGTTTACTGCATGCCTGGAGAGAACACAGCAGTAAAAACCAGGTTTGCTACTGGAAAAAGAGGAAAGAGAAGACTTTCATTGACGGACCCAGCCATGGCAGCGTAGCAGCCCTGCGTTTTAGACGGCAGCAGCTCGGGACTCTGGACGTGTGTTTGCCCTCAAGTTTGCTAAGCTGCTGGTTTATTACTGAAGAAAGAATGTGGCAGATTGTTTTCTTTACTCTGAGCTGTGATCTTGTCTTGGCCGCAGCCTATAACAACTTTCGGAAGAGCATGGACAGCATAGGAAAGAAGCAATATCAGGTCCAGCATGGGTCCTGCAGCTACACTTTCCTCCTGCCAGAGATGGACAACTGCCGCTCTTCCTCCAGCCCCTACGTGTCCAATGCTGTGCAGAGGGACGCGCCGCTCGAATACGATGACTCGGTGCAGAGGCTGCAAGTGCTGGAGAACATCATGGAAAACAACACTCAGTGGCTAATGAAG
Seq A exon
CTTGAGAATTATATCCAGGACAACATGAAGAAAGAAATGGTAGAGATACAGCAGAATGCAGTACAGAACCAGACGGCTGTGATGATAGAAATAGGGACAAACCTGTTGAACCAAACAGCGGAGCAAACGCGGAAGTTAACTGATGTGGAAGCCCAA
Seq C2 exon
GTATTAAATCAGACCACGAGACTTGAACTTCAGCTCTTGGAACACTCCCTCTCGACAAACAAATTGGAAAAACAGATTTTGGACCAGACCAGTGAAATAAACAAATTGCAAGATAAGAACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000091879-'0-2,'0-1,2-2
Average complexity
S
Mappability confidence:
100%=100=100%
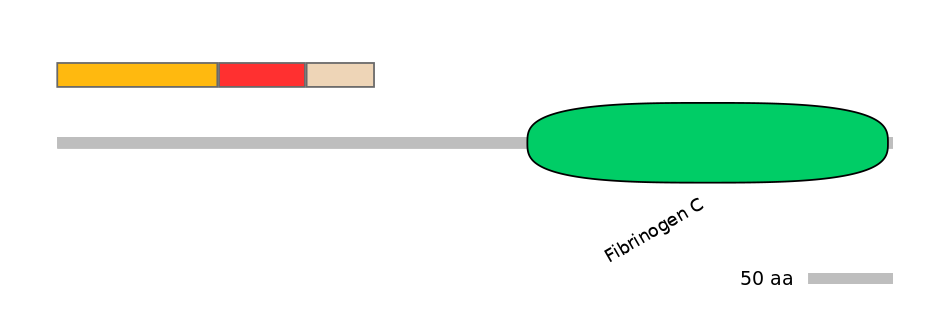
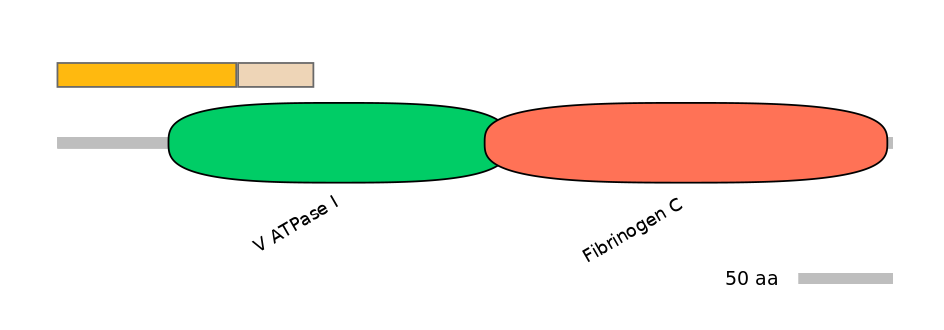
Protein Impact
Alternative protein isoforms (Ref)
Show structural model
Features
Disorder rate (Iupred):
C1=0.010 A=0.096 C2=0.000
Domain overlap (PFAM):
C1:
PF0149614=V_ATPase_I=PU(19.8=37.5)
A:
NO
C2:
PF0149614=V_ATPase_I=FE(22.0=100),PF0497710=DivIC=PU(60.0=73.2)
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TATCAGGTCCAGCATGGGTCC
R:
TGTCGAGAGGGAGTGTTCCAA
Band lengths:
246-402
Functional annotations
There are 2 annotated functions for this event
PMID: 10766762
This event
[Negative results]. Angiopoietin-2 (Ang-2) is a naturally occurring antagonist of angiopoietin-1 (Ang1) that competes for binding to the Tie2 receptor and blocks Ang1-induced Tie2 autophosphorylation during vasculogenesis. Using the polymerase chain reaction, the authors isolated a cDNA encoding a novel shorter form of Ang2 from human umbilical vein endothelial cell cDNA and have designated it angiopoietin-2_443 (Ang2_443), because it contains 443 amino acids (lacks exon 2 (HsaEX0004099)). Part of the coiled-coil domain (amino acids 96?148) is absent in Ang2_443 because of alternative splicing of the gene. Like Ang2, recombinant Ang2_443 expressed in COS-7 cells is secreted as a glycosylated homodimeric protein. Recombinant Ang2443 binds to the Tie2 receptor but does not induce Tie2 phosphorylation. Pre-occupation of Ang2443 on Tie2 inhibits Ang1 or Ang2 binding and inhibits Ang1-induced phosphorylation.
PMID: 33037065
The authors studied the function of ANGPT2443 in cellular experiments as well as in a genetic model in vivo, revealing that it is proteolytically cleaved into a lower molecular weight isoform (termed ANGPT2DAP) that lacks the superclustering domain necessary for multimer formation. When compared with full-length ANGPT2, ANGPT2443 and ANGPT2DAP showed lower binding affinity to alpha5beta1 integrin, but were more potent inhibitors of ANGPT1-TIE2 signaling. Functionally, ANGPT2443 impaired vessel enlargement and vein morphogenesis during postnatal retinal angiogenesis. Tumor experiments in Angpt2443-expressing mice showed enhanced destabilization of the lung vasculature, with varying effects on metastasis. Taken together, the study provides important insight into the significance of ANGPT2 alternative splicing and identifies ANGPT2443 and ANGPT2DAP as a biological rheostat of ANGPT1-TIE2 signaling. Future work will need to characterize the relative ratios and functional contributions of the ANGPT2 variants in different pathophysiologic settings.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)