HsaEX0020455 @ hg38
Exon Skipping
Gene
ENSG00000079805 | DNM2
Description
dynamin 2 [Source:HGNC Symbol;Acc:HGNC:2974]
Coordinates
chr19:10805916-10812377:+
Coord C1 exon
chr19:10805916-10805967
Coord A exon
chr19:10808569-10808580
Coord C2 exon
chr19:10812264-10812377
Length
12 bp
Sequences
Splice sites
3' ss Seq
CTAACTTTGCATATTCAAAGGGG
3' ss Score
3.62
5' ss Seq
CTGGTAAGT
5' ss Score
10.65
Exon sequences
Seq C1 exon
TGCCCAGCAGAGGAGCACGCAGCTGAACAAGAAGAGAGCCATCCCCAATCAG
Seq A exon
GGGGAGATCCTG
Seq C2 exon
GTGATCCGCAGGGGCTGGCTGACCATCAACAACATCAGCCTGATGAAAGGCGGCTCCAAGGAGTACTGGTTTGTGCTGACTGCCGAGTCACTGTCCTGGTACAAGGATGAGGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000079805-'56-53,'56-47,58-53
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.071 A=0.000 C2=0.000
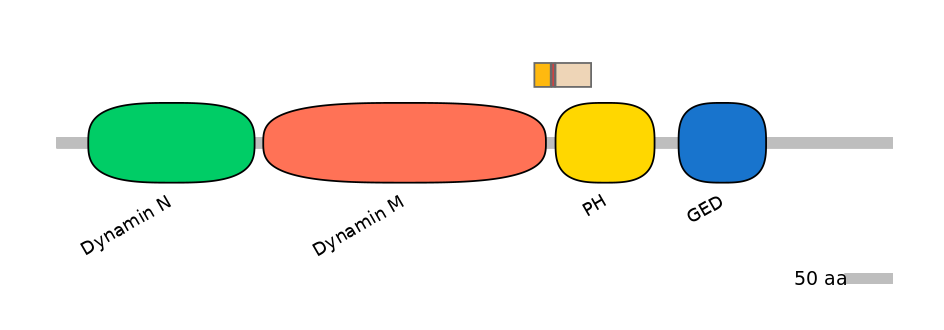
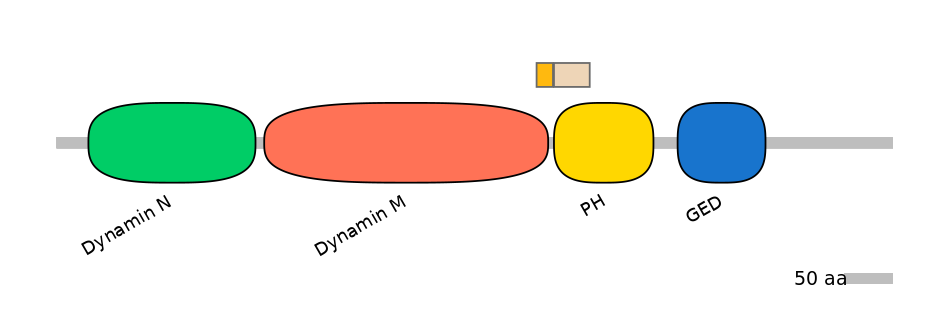
Domain overlap (PFAM):
C1:
PF0103115=Dynamin_M=PD(4.1=66.7)
A:
NO
C2:
PF0016924=PH=PU(35.6=97.4)
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACAAGAAGAGAGCCATCCCCA
R:
AACCAGTACTCCTTGGAGCCG
Band lengths:
97-109
Functional annotations
There are 4 annotated functions for this event
PMID: 20127478
[Review conclusion]. Altogether, these data suggest a preferential involvement of isoforms 1 and 3 in clathrin and caveolae-dependent endocytosis, whereas isoforms 2 and 4 participate in uncoated endocytosis and trafficking from the Golgi apparatus.
PMID: 9725914
The authors generated stably transfected Clone 9 cells expressing full-length cDNAs of six different spliced forms of the three dynamin genes (Dnm1, Dnm2, and Dnm3) tagged with green fluorescent protein. The investigated splice variants are: Dnm1 (with mutually exclusive exon 10a (MmuEX6100535) or exon 10b (MmuEX0015279), Dnm2 (with/without cassette exon (MmuEX0015304), and Dnm3 (with/without cassette exon MmuEX0015311). Confocal or fluorescence microscopy of these transfected cells revealed that many of the dynamin proteins associate with distinct membrane compartments, which include clathrin-coated pits at the plasma membrane and the Golgi apparatus, and several undefined vesicle populations.
PMID: 10780938
The authors expressed each of the four dynamin 2 isoforms (aa, ab, ba, and bb) as either wild-type proteins or GTPase-defective mutants. When expressed as enhanced green fluorescent protein (EGFP) fusions, these isoforms were found to have overlapping subcellular distributions being localized throughout the cell cytoplasm, on punctate vesicles and in a perinuclear compartment. This distribution was qualitatively similar to that of endogenous dynamin 2 and overlapped with GLUT-4 in the basal state. Expression of wild-type dynamin 2 isoforms had no effect on the basal or insulin-stimulated distribution of GLUT-4; however, expression of the dominant-interfering dynamin 2 mutants similarly inhibited GLUT-4 endocytosis. These data demonstrate that dynamin 2 is required for GLUT-4 endocytosis in 3T3L1 adipocytes and suggest that, relative to GLUT-4 trafficking, the dynamin 2 splice variants have overlapping functions and are probably not responsible for mediating distinct GLUT-4 budding events.
PMID: 15811383
In vitro binding studies using two Dyn2 spliced forms (aa and bb) and Cav1 peptides immobilized on paper identify specific domains of Cav1 that bind Dyn2. Interestingly, these Cav1-binding domains differ markedly between two spliced variant forms of Dyn2. In a hepatocyte cell line, the K44A-isoform 1 inhibits caveolae-dependent internalization, but not the other K44A mutant isoforms. Isoform 1 (aa): Includes MmuEX0015299+MmuEX0015304. Isoform 2 (ba): MmuEX0015305+MmuEX0015304. Isoform 3 (ab): MmuEX0015299. Isoform 4 (bb): MmuEX0015305.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development