HsaEX0033123 @ hg38
Exon Skipping
Gene
ENSG00000162687 | KCNT2
Description
potassium sodium-activated channel subfamily T member 2 [Source:HGNC Symbol;Acc:HGNC:18866]
Coordinates
chr1:196342079-196373248:-
Coord C1 exon
chr1:196373140-196373248
Coord A exon
chr1:196343056-196343105
Coord C2 exon
chr1:196342079-196342228
Length
50 bp
Sequences
Splice sites
3' ss Seq
CAGAAATTCATTTCTTACAGTTC
3' ss Score
5.44
5' ss Seq
CTGGTGAGG
5' ss Score
8.3
Exon sequences
Seq C1 exon
ATCATGTTGTTTGTGAAGAAGAGTTTAAATACGCCATGTTAGCTTTAAACTGTATATGCCCAGCAACATCTACACTTATTACACTACTGGTTCATACCTCTAGAGGGCA
Seq A exon
TTCTGAAGGCTGGAAATCCAAGATCTAGGTGCCAACATGATCCAGTGCTG
Seq C2 exon
AGAAGGCCAGCAATCGCCAGAACAATGGCAGAAGATGTACGGTAGATGCTCCGGGAATGAAGTCTACCACATTGTTTTGGAAGAAAGTACATTTTTTGCTGAATATGAAGGAAAGAGTTTTACATATGCCTCTTTCCATGCACACAAAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000162687_MULTIEX1-3/4=1-4
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
Show structural model
Features
Disorder rate (Iupred):
C1=0.000 A=0.056 C2=0.057
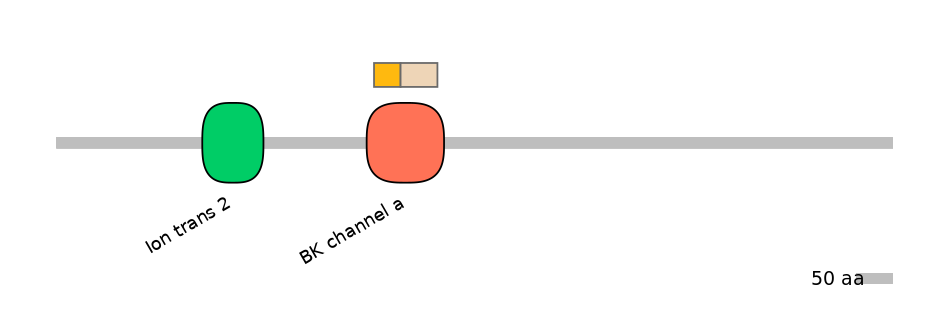
Domain overlap (PFAM):
C1:
PF0349313=BK_channel_a=FE(34.0=100)
A:
NO
C2:
PF0349313=BK_channel_a=FE(47.2=100)
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACGCCATGTTAGCTTTAAACTGT
R:
TTCCCGGAGCATCTACCGTAC
Band lengths:
136-186
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development