HsaEX0037512 @ hg19
Exon Skipping
Gene
ENSG00000076984 | MAP2K7
Description
mitogen-activated protein kinase kinase 7 [Source:HGNC Symbol;Acc:6847]
Coordinates
chr19:7968776-7974781:+
Coord C1 exon
chr19:7968776-7968953
Coord A exon
chr19:7970693-7970740
Coord C2 exon
chr19:7974640-7974781
Length
48 bp
Sequences
Splice sites
3' ss Seq
TCTTCCTTTTCCCCATCCAGTTA
3' ss Score
9.41
5' ss Seq
CAGGTACCA
5' ss Score
7.88
Exon sequences
Seq C1 exon
CTGCCGGACTGACGGGCGGCCGGGCGGTGCGCGGCGGCGGTGGCGGCGGGGAAGATGGCGGCGTCCTCCCTGGAACAGAAGCTGTCCCGCCTGGAAGCAAAGCTGAAGCAGGAGAACCGGGAGGCCCGGCGGAGGATCGACCTCAACCTGGATATCAGCCCCCAGCGGCCCAGGCCCA
Seq A exon
TTATTGTGATCACTCTAAGCCCTGCTCCTGCCCCGTCCCAACGAGCAG
Seq C2 exon
CCCTGCAGCTCCCGCTGGCCAACGATGGGGGCAGCCGCTCGCCATCCTCAGAGAGCTCCCCGCAGCACCCCACGCCCCCCGCCCGGCCCCGCCACATGCTGGGGCTCCCGTCAACCCTGTTCACACCCCGCAGCATGGAGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000076984_MULTIEX1-2/2=C1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.981 A=0.765 C2=0.892
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
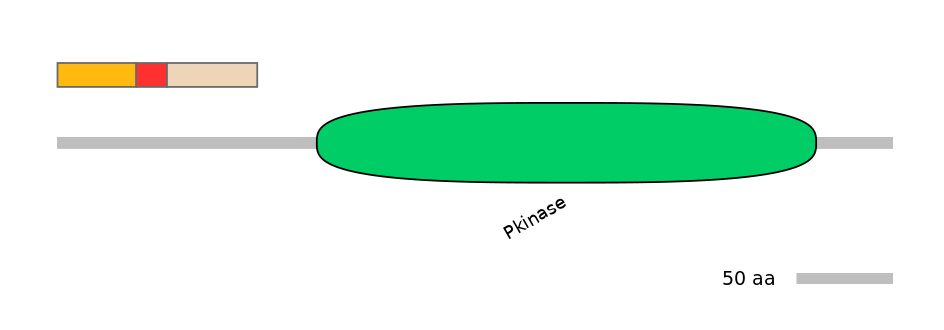
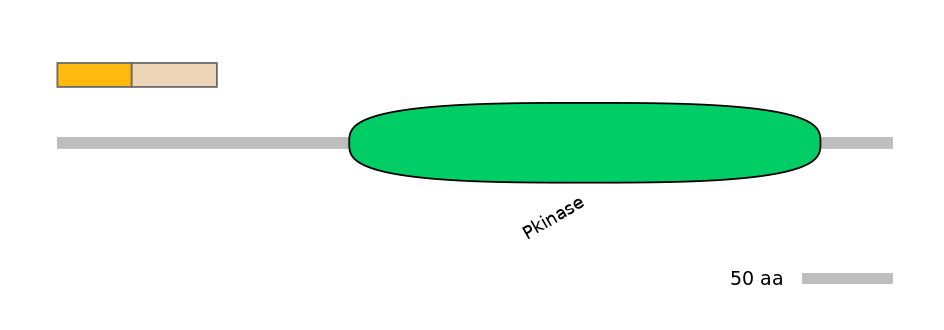
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AAAGCTGAAGCAGGAGAACCG
R:
GAGCTCTCTGAGGATGGCGAG
Band lengths:
138-186
Functional annotations
There are 3 annotated functions for this event
PMID: 16533805
This event
MAP2K7 recognizes its target JNK by a novel mechanism involving a partially cooperative interaction of three low affinity D-sites in the N-terminal domain of MAP2K7, in the alternative exon. Mutations of the conserved residues within any one of the three docking sites (D1, D2, and D3) disrupted the ability of the N-terminal domain of MAP2K7beta to bind JNK1 by about 50-70%. Moreover, mutation of any two of the three D-sites reduced binding by about 80-90%, and mutation of all three reduced binding by 95%.
PMID: 26443849
This event
In response to T-cell activation, the Jun kinase (JNK) kinase MAP kinase kinase 7 (MKK7, MAP2K7) is alternatively spliced to favor an isoform that lacks exon 2. This isoform restores a JNK-docking site within MAP2K7 that is disrupted in the larger isoform. Consistently, skipping of MAP2K7 exon 2 enhances JNK pathway activity, as indicated by c-Jun phosphorylation and up-regulation of TNF-alpha. Moreover, this splicing event is itself dependent on JNK signaling. Thus, MAP2K7 alternative splicing represents a positive feedback loop through which JNK promotes its own signaling. Repression of MAP2K7 exon 2 is dependent on the presence of flanking sequences and the JNK-induced expression of the RNA-binding protein CELF2, which binds to these regulatory elements. Finally, _25% of T-cell receptor-mediated alternative splicing events are dependent on JNK signaling. These JNK-dependent events are also significantly enriched for responsiveness to CELF2.
PMID: 32601196
This event
Down-regulation of MBNL1 predicted poor overall survival in breast, lung, and stomach adenocarcinomas and increased relapse and distant metastasis in triple-negative breast cancer. A discrete set of alternative splicing events (ASEs) are shared between MBNL1-low cancers and embryonic stem cells including a MAP2K7 delta-exon2 splice variant that leads to increased stem/progenitor-like properties via JNK activation. Accordingly, JNK inhibition is capable of reversing MAP2K7 delta-exon2-driven tumor dedifferentiation in MBNL1-low cancer cells. To directly examine the role of MAP2K7 delta-exon2 splice isoform in cellular dedifferentiation and cancer stem/progenitor-like properties, the authors used an antisense morpholino (AMO) directed against the 5_ splice site of MAP2K7 exon2 to force MAP2K7 exon2 skipping in HFE-145 cells. This produced robust exon2 skipping and activation of JNK signaling as previously reported. Most importantly it drove up-regulation of CSC and pluripotency markers (CD133, NANOG, and SOX2) and increased trans-well migration and invasion, thereby recapitulating stemness phenotype observed upon loss of MBNL1 expression. Surprisingly, the MAP2K7 AMO also led to up-regulation of MAP2K7 and a modest (_30%) down-regulation of MBNL1 proteins, suggesting feedback signaling.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)