HsaEX0053093 @ hg19
Exon Skipping
Gene
ENSG00000084093 | REST
Description
RE1-silencing transcription factor [Source:HGNC Symbol;Acc:9966]
Coordinates
chr4:57785953-57802010:+
Coord C1 exon
chr4:57785953-57786036
Coord A exon
chr4:57793781-57793830
Coord C2 exon
chr4:57796007-57802010
Length
50 bp
Sequences
Splice sites
3' ss Seq
TATGCATTCCATTGGACCAGTGG
3' ss Score
3.73
5' ss Seq
TGGGTATGT
5' ss Score
7.43
Exon sequences
Seq C1 exon
GAGAACGCCCATATAAATGTGAACTTTGTCCTTACTCAAGTTCTCAGAAGACTCATCTAACTAGACATATGCGTACTCATTCAG
Seq A exon
TGGGGTATGGATACCATTTGGTAATATTTACTAGAGTGTGATCTAGATGG
Seq C2 exon
GTGAGAAGCCATTTAAATGTGATCAGTGCAGTTATGTGGCCTCTAATCAACATGAAGTAACCCGCCATGCAAGACAGGTTCACAATGGGCCTAAACCTCTTAATTGCCCACACTGTGATTACAAAACAGCAGATAGAAGCAACTTCAAAAAACATGTAGAGCTACATGTGAACCCACGGCAGTTCAATTGCCCTGTATGTGACTATGCAGCTTCCAAGAAGTGTAATCTACAGTATCACTTCAAATCTAAGCATCCTACTTGTCCTAATAAAACAATGGATGTCTCAAAAGTGAAACTAAAGAAAACCAAAAAACGAGAGGCTGACTTGCCTGATAATATTACCAATGAAAAAACAGAAATAGAACAAACAAAAATAAAAGGGGATGTGGCTGGAAAGAAAAATGAAAAGTCCGTCAAAGCAGAGAAAAGAGATGTCTCAAAAGAGAAAAAGCCTTCTAATAATGTGTCAGTGATCCAGGTGACTACCAGAACTCGAAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000084093-'8-9,'8-6,9-9
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.069 A=0.000 C2=0.823
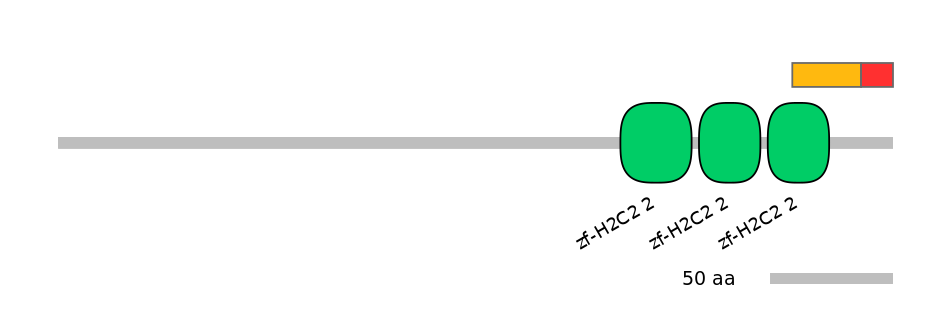
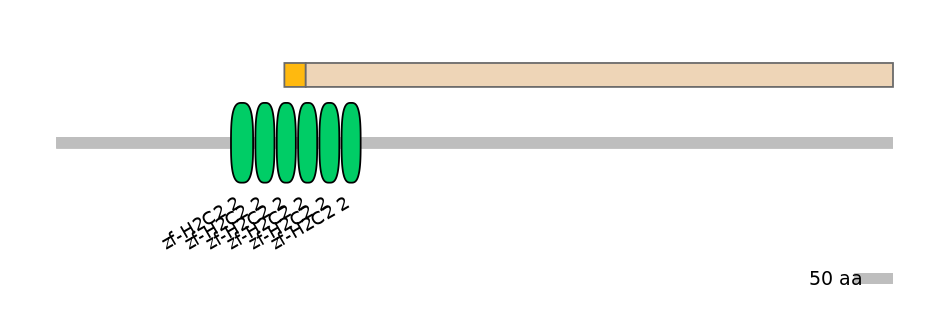
Domain overlap (PFAM):
C1:
PF134651=zf-H2C2_2=PD(57.7=51.7),PF134651=zf-H2C2_2=PU(38.5=34.5)
A:
NO
C2:
PF134651=zf-H2C2_2=PD(57.7=1.9),PF134651=zf-H2C2_2=WD(100=3.5),PF134651=zf-H2C2_2=WD(100=3.4)
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGTGAACTTTGTCCTTACTCAAGT
R:
TCTTGCATGGCGGGTTACTTC
Band lengths:
141-191
Functional annotations
There are 4 annotated functions for this event
PMID: 29961578
This event
The absence of this exon in transcripts is responsible for one form of dominant hereditary deafness in human patients and mouse model systems. Inclusion of this exon in the REST transcript provides post-translational inactivation of the REST protein, which is critical for the survival of mechanosensory hair cells in the ear. This exon is essential for the derepression of many neuronal genes in hair cells; without this exon deafness occurs.
PMID: 10490617
Inclusion form has a PTC. It lacks DNA-binding capability, acts as a dominant negative protein which sequesters full-length REST in nonfunctional hetero-oligomers.
PMID: 21884984
Cross regulation with nSR100, which enhances the inclusion of the alternative exon in mouse
PMID: 30995482
Splicing alterations in the Srrm3/Srrm4 DKO mice prevent inactivation of the gene repressor REST, which maintains immature excitatory GABAergic neurotransmission by repressing K-Cl cotransporter 2. Low REST activity is also due to a (likely indirect) reduction in REST expression levels.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)