GgaEX0017156 @ galGal4
Exon Skipping
Gene
ENSGALG00000011390 | REST
Description
RE1-silencing transcription factor [Source:HGNC Symbol;Acc:HGNC:9966]
Coordinates
chr4:48626470-48632680:-
Coord C1 exon
chr4:48632597-48632680
Coord A exon
chr4:48628479-48628527
Coord C2 exon
chr4:48626470-48628166
Length
49 bp
Sequences
Splice sites
3' ss Seq
CAATGCATTCAATGGACCAGTTG
3' ss Score
0.39
5' ss Seq
CCGGTATGT
5' ss Score
9.2
Exon sequences
Seq C1 exon
GAGAGCGACCCTATCAATGTGCTATGTGTCCCTATTCCAGCTCTCAGAAGACCCATTTAACCAGGCACATGCGCACCCACTCAG
Seq A exon
TTGGGTATGGATTCCATTTGCTAATAGTACTGTAGAGAGATCTCTGCCG
Seq C2 exon
GTGAGAAGCCATTCAAATGTGATCAGTGCAGTTACGTGGCCTCAAACCAGCATGAAGTAACTCGTCATGCAAGGCAAGTTCACAATGGGCCGAAGCCTCTGACTTGCCCGCACTGTGACTACAAAACAGCCGATCGCAGCAATTTCAAAAAGCACGTTGAGCTCCACGTCAATCCGCGCCAGTTCCTTTGTCCTGTTTGTGACTACGCGGCATCTAAGAAGTGTAACCTGCAGTATCACATCAAATCCAGGCATCCCGATTGTTCGGACATCACCATGGATGTTTCAAAGGTGAAGCTACGGACTAAAAAGAGCGAAGCTGACTTTTCCGAGAGCATCAGTGACAAAGTGGAGAAAGAGCAAACAAAAGGGGATTCGCCCACAAAGAAAACTGAGAAAACTGTGAAAGTGGAGAAAAAAGAGAACTTGGCAAAGGAAAAGAAGCCAACGAGCAATGTTCCTGCACGTCAGGTTACGACCAGAAGTCGGAAATCGACTTCA
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000011390_MULTIEX1-2/2=1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion (Alt. Stop)
Show structural model
Features
Disorder rate (Iupred):
C1=0.069 A=0.000 C2=0.765
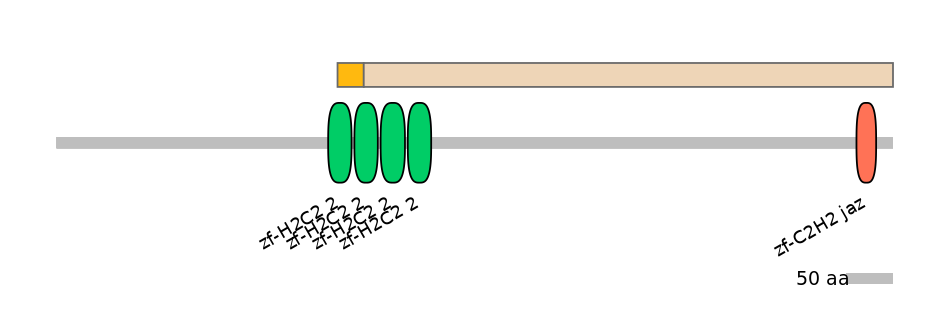
Domain overlap (PFAM):
C1:
PF134651=zf-H2C2_2=PD(57.7=51.7),PF134651=zf-H2C2_2=PU(38.5=34.5)
A:
NO
C2:
PF134651=zf-H2C2_2=PD(57.7=2.7),PF134651=zf-H2C2_2=WD(100=4.8),PF134651=zf-H2C2_2=WD(100=4.6),PF121713=zf-C2H2_jaz=WD(100=3.9)
Main Inclusion Isoform:
ENSGALT00000018580fB2011
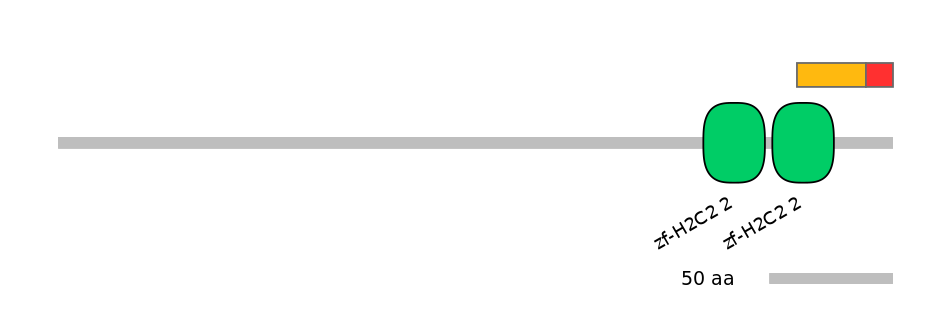
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTCCAGCTCTCAGAAGACCCA
R:
TGAACTTGCCTTGCATGACGA
Band lengths:
132-181
Functional annotations
There are 4 annotated functions for this event
PMID: 29961578
The absence of this exon in transcripts is responsible for one form of dominant hereditary deafness in human patients and mouse model systems. Inclusion of this exon in the REST transcript provides post-translational inactivation of the REST protein, which is critical for the survival of mechanosensory hair cells in the ear. This exon is essential for the derepression of many neuronal genes in hair cells; without this exon deafness occurs.
PMID: 10490617
Inclusion form has a PTC. It lacks DNA-binding capability, acts as a dominant negative protein which sequesters full-length REST in nonfunctional hetero-oligomers.
PMID: 21884984
Cross regulation with nSR100, which enhances the inclusion of the alternative exon in mouse
PMID: 30995482
Splicing alterations in the Srrm3/Srrm4 DKO mice prevent inactivation of the gene repressor REST, which maintains immature excitatory GABAergic neurotransmission by repressing K-Cl cotransporter 2. Low REST activity is also due to a (likely indirect) reduction in REST expression levels.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]