HsaEX0065118 @ hg38
Exon Skipping
Gene
ENSG00000146872 | TLK2
Description
tousled like kinase 2 [Source:HGNC Symbol;Acc:HGNC:11842]
Coordinates
chr17:62596585-62600820:+
Coord C1 exon
chr17:62596585-62596674
Coord A exon
chr17:62597220-62597258
Coord C2 exon
chr17:62600651-62600820
Length
39 bp
Sequences
Splice sites
3' ss Seq
TGTCTGTTTTATCTTTATAGATT
3' ss Score
10.79
5' ss Seq
ATAGTATGT
5' ss Score
4.41
Exon sequences
Seq C1 exon
GCATGCATGTAGGGAATACCGGATTCATAAAGAGCTGGATCATCCCAGAATAGTTAAGCTGTATGATTACTTTTCACTGGATACTGACTC
Seq A exon
ATTTCCTATTCTGGACCCTCATATGCATGGAATCATATA
Seq C2 exon
GTTTTGTACAGTATTAGAATACTGTGAGGGAAATGATCTGGACTTCTACCTGAAACAGCACAAATTAATGTCGGAGAAAGAGGCCCGGTCCATTATCATGCAGATTGTGAATGCTTTAAAGTACTTAAATGAAATAAAACCTCCCATCATACACTATGACCTCAAACCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000146872_MULTIEX2-18/22=17-19
Average complexity
C2
Mappability confidence:
99%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
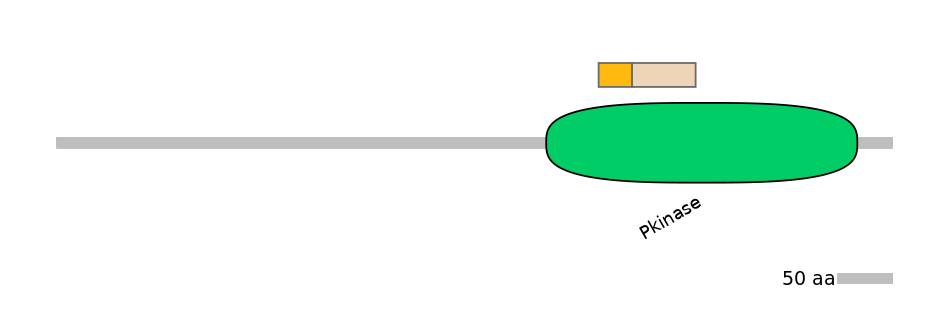
Domain overlap (PFAM):
C1:
PF0006920=Pkinase=FE(10.7=100)
A:
NA
C2:
PF0006920=Pkinase=FE(20.4=100)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACCGGATTCATAAAGAGCTGGA
R:
GTCCAGATCATTTCCCTCACAGT
Band lengths:
116-155
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development