HsaEX0073616 @ hg19
Exon Skipping
Gene
ENSG00000170954 | ZNF415
Description
zinc finger protein 415 [Source:HGNC Symbol;Acc:20636]
Coordinates
chr19:53611133-53619686:-
Coord C1 exon
chr19:53619566-53619686
Coord A exon
chr19:53618463-53618560
Coord C2 exon
chr19:53611133-53613161
Length
98 bp
Sequences
Splice sites
3' ss Seq
GTTTCGCTCTTATTGCCCAGGCT
3' ss Score
6.39
5' ss Seq
CAGGTATGC
5' ss Score
9.37
Exon sequences
Seq C1 exon
TTGACATTCAGGGACGTGGCCATCGAATTCTCTCAAGATGAGTGGAAATGCCTGAACTCTACACAGAGGACTTTATACAGGGATGTGATGTTGGAGAACTACAGGAACCTGGTCTCCCTGG
Seq A exon
GCTGGAGTGCAATGGTGCAATCTCGGCTCACTGCAACCTTCGCCTCCCGGATTCAAATGATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACAG
Seq C2 exon
ATCTGTCTCGTAACTGTGTAATCAAGGAACTAGCACCACAACAGGAAGGTAACCCAGGAGAAGTATTCCACACAGTGACATTGGAACAACATGAAAAACATGACATTGAAGAGTTTTGCTTCAGGGAAATCAAGAAAAAAATACACGACTTTGACTGTCAGTGGAGAGATGATGAAAGAAATTGCAACAAAGTGACTACGGCCCCAAAAGAAAATCTTACTTGTAGGAGAGACCAACGCGATAGAAGAGGTATAGGAAACAAGTCTATTAAACATCAGCTTGGATTAAGCTTTCTACCACATCCCCATGAACTGCAGCAGTTTCAAGCTGAAGGGAAAATTTATGAATGTAACCATGTTGAGAAGTCTGTCAACCATGGTTCCTCAGTTTCACCACCCCAAATAATTTCTTCTACCATCAAAACCCATGTTTCTAATAAATATGGGACTGATTTCATCTGTTCTTCATTACTCACACAAGAACAGAAATCATGCATTAGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000170954_MULTIEX1-5/5=3-C2
Average complexity
C2*
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
Show structural model
Features
Disorder rate (Iupred):
C1=0.013 A=0.275 C2=0.108
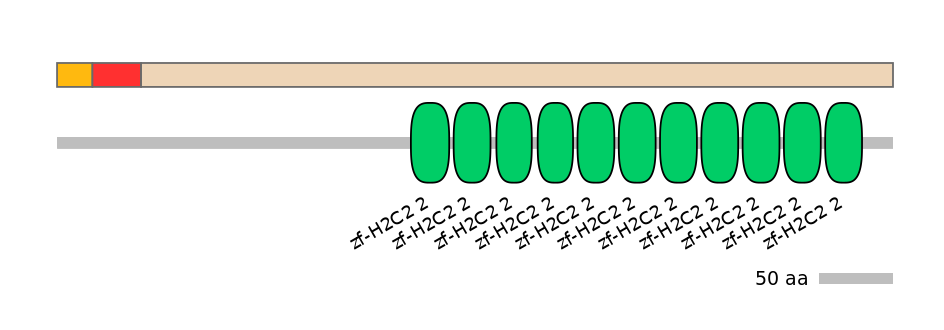
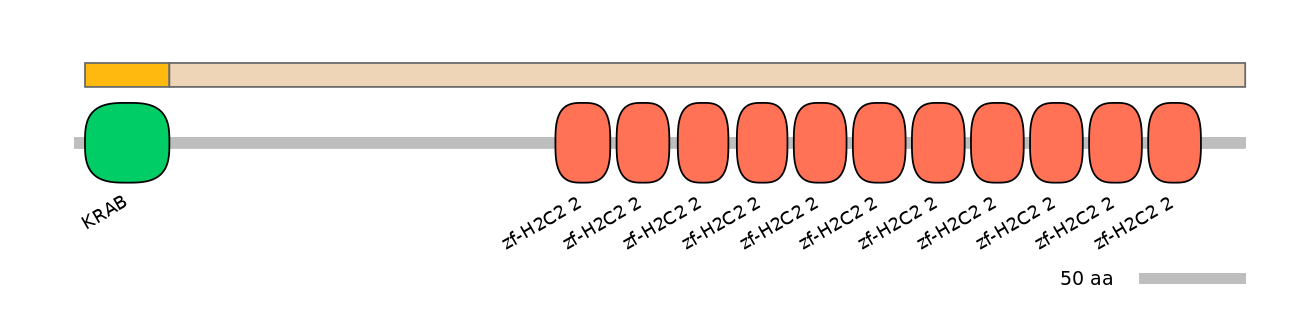
Domain overlap (PFAM):
C1:
PF0135222=KRAB=WD(100=100.0)
A:
PF0135222=KRAB=PD(0.1=0.0)
C2:
PF134651=zf-H2C2_2=WD(100=5.3),PF134651=zf-H2C2_2=WD(100=5.1),PF0013017=C1_1=WD(100=12.5),PF134651=zf-H2C2_2=WD(100=4.9),PF134651=zf-H2C2_2=WD(100=4.9),PF134651=zf-H2C2_2=WD(100=5.1),PF134651=zf-H2C2_2=WD(100=5.1),PF134651=zf-H2C2_2=WD(100=5.1),PF0013017=C1_1=WD(100=10.6),PF134651=zf-H2C2_2=WD(100=5.1),PF134651=zf-H2C2_2=WD(100=5.1),PF134651=zf-H2C2_2=WD(100=5.1),PF0013017=C1_1=WD(100=11.7),PF134651=zf-H2C2_2=WD(100=5.1)
Other Inclusion Isoforms:
Associated events
Conservation
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGACGTGGCCATCGAATTCTC
R:
TCCTGGGTTACCTTCCTGTTGT
Band lengths:
169-267
Functional annotations
There are 1 annotated functions for this event
PMID: 17055453
This event
Isoforms 2-5 are cytoplasmic, isoform 1 is nuclear. Isoforms 2-5, but not 1, inhibit AP-1 and p53. Isoforms are all expressed in both adult and fetal tissue. Exon 3 = HsaEX0073614, Exon 4 = HsaEX0073615, ALTA in Exon 4 = HsaALTA0009817, Exon 5 = HsaEX0073616. Isoform 1: skips exons 3, 4 and 5; Isoform 2: skips exon 4; Isoform 3: short version of exon 4; Isoform 4: long form of exon 4 and skipping of exon 5; Isoform 5: skippin of exons 4 and 5.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)