HsaEX1045137 @ hg38
Exon Skipping
Gene
ENSG00000185177 | ZNF479
Description
zinc finger protein 479 [Source:HGNC Symbol;Acc:HGNC:23258]
Coordinates
chr7:57126592-57128931:-
Coord C1 exon
chr7:57128852-57128931
Coord A exon
chr7:57127517-57127596
Coord C2 exon
chr7:57126592-57126718
Length
80 bp
Sequences
Splice sites
3' ss Seq
GCTCTTATCTTTATTTACAGGTG
3' ss Score
10.31
5' ss Seq
TTGGTGAGC
5' ss Score
7.43
Exon sequences
Seq C1 exon
GTTTGATATGTCCAGAGCATCTCATCTGAAAATACATTTCAGAGAAAGAGGAGGAAAAGAAACAAATCACTTTTTCTCAG
Seq A exon
GTGAGAGAAACTGGGAAAAACCCAGACTCTGCCATTTACTGGATATTTGACAAAATATTCTTACTAGGCTAGAAACATTG
Seq C2 exon
GGACTGTTGACATTCAGAGACATAGCTATAGAATTCTCTCTGGAGGAATGGCAATGCCTGGATTGTGCTCAGCGGAATTTATATAGAGATGTGATGTTAGAGAACTACAGAAACCTGGTCTCCCTGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000185177-'5-10,'5-9,6-10
Average complexity
C3
Mappability confidence:
88%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.174 C2=0.000
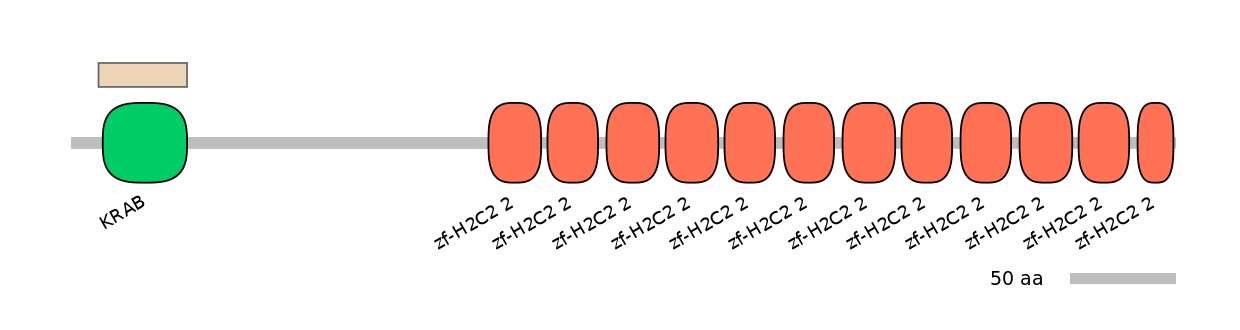
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
PF0135222=KRAB=WD(100=95.3)
Main Inclusion Isoform:
ENST00000620639fB30768
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACATTTCAGAGAAAGAGGAGGAA
R:
GGAGACCAGGTTTCTGTAGTTCT
Band lengths:
170-250
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains