MmuEX0022277 @ mm10
Exon Skipping
Gene
ENSMUSG00000016283 | H2-M2
Description
histocompatibility 2, M region locus 2 [Source:MGI Symbol;Acc:MGI:95914]
Coordinates
chr17:37480851-37483246:-
Coord C1 exon
chr17:37482977-37483246
Coord A exon
chr17:37482483-37482758
Coord C2 exon
chr17:37480851-37481747
Length
276 bp
Sequences
Splice sites
3' ss Seq
AACTTGTGATACGGAACCAGGTT
3' ss Score
1.23
5' ss Seq
CAGGTACGA
5' ss Score
10.03
Exon sequences
Seq C1 exon
GATCCCACTCCTTGCGGTACTTCGACATCGCAGTGTCAAGACCTGGCCTAGAGGAGACCCACTACATGACTGTTGGCTATGTGGATGACACAGAGTTTGTGCATTTTGACAATGAGGCTGAGAATCCGAGGTTTGAGCCCCGAGTGCCTTGGATGGAACAGATGGGACAGAAGTACTGGGATGACCAGACACGCATTGCGAAAGCTGCAGAACAGCAGATTAGAGTGTACTTTCAGAAACTGCGAGACTACTACAACCAGAGCCAGAACA
Seq A exon
GTTCTCACACCATCCAAAGGATGACTGGCTGCTACATTGGACCAGACGGGCACCTACTTCACGCATACCGTCAGTTTGGTTATGATGGGCAAGATTACCTCACTCTGAATGAAGATCTGAGCACCTGGACTGCAGCAGATGCGGCAGCTGAGATCACCCGTAGAGAGTGGGAGGCAACTAATGTGGCTGAGTTCTGGAGGGTCTACTTGGAGGGCCCTTGCATGGTGTGGCTCTTTAAATACCTGACAGTGGGAAATGAGACTCTACTGCGCACAG
Seq C2 exon
AGCCCCCAAAGGCATATGTGACCCATCACCCCAGACCTGAAGGTGATGTCACCCTGAGGTGCTGGGCCCTGGGTTTCTACCCTTCTGACATCATCATGATCTGGCAAAGAGATGGGGAGGACCAGACCCAGGACATGGATGTTATTGAGACCAGACCTGCAGGGGATGGAACCTTCCAGAAGTGGGTAGCTGTGGTGGTGCCTTCTGGGAAGGAGCAGAATTACACATGCCATGTGGCTCATGAGGGACTGCCTGAGCCCCTCACCCTGAGATGGAGTAAGTAGGATGTAGGCACATAGCCTGTGGTTAGAGAAGGCAAAAGCCTTTCTGGAGATCCTGAGCAGGGTCAAGGTTGAGATCTGGGGTCAGGGTCCTACAATTTTACATTTCTTTTCAGGTAGACCTCCTCAGTCTTTCATTTTCATCATAATAGTTGCTGTTGGCCTGGTTCTCCTGGGAGCTTCAGTGGCTACTCTTGTCATGTGGAAGAAGAGCTCGGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000016283-'1-3,'1-1,2-3
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
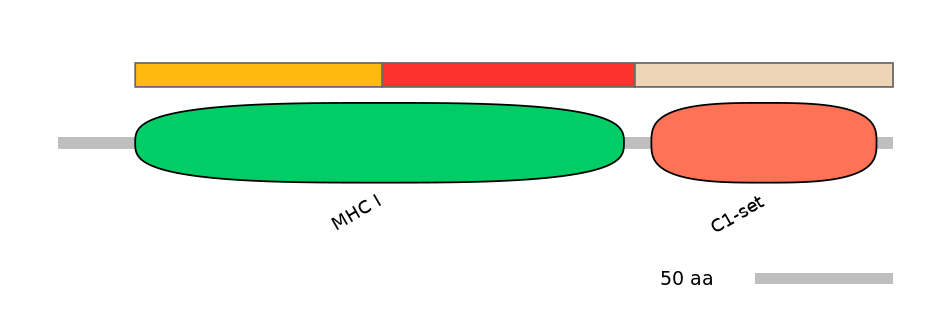
Features
Disorder rate (Iupred):
C1=0.022 A=0.000 C2=0.157
Domain overlap (PFAM):
C1:
PF0012913=MHC_I=PU(50.3=98.9)
A:
PF0012913=MHC_I=PD(49.2=94.6)
C2:
PF0765410=C1-set=WD(100=87.4)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATCCCACTCCTTGCGGTACTT
R:
GGTCACATATGCCTTTGGGGG
Band lengths:
292-568
Functional annotations
There are 2 annotated functions for this event
PMID: 11290782
Four membrane-bound (HLA-G1 to -G4) and two soluble (HLA-G5, and -G6) proteins are generated by alternative splicing. Only HLA-G1 has been extensively studied in terms of both expression and function. The authors provide evidence here that HLA-G2, -G3, and -G4 truncated isoforms reach the cell surface of transfected cells, as endoglycosidase H-sensitive glycoproteins, after a 2-h chase period. Moreover, cytotoxicity experiments show that these transfected cells are protected from the lytic activity of both innate (NK cells) and acquired (CTL) effectors, although to slightly different levels. The full-length transcript (HLA-G1) encodes three extracellular domains (alpha1, alpha2, and alpha3), a transmembrane region, and a cytoplasmic tail. Three other spliced transcripts encode membrane-bound proteins that lack alpha2 (HLA-G2; skipping exon 3, HsaEX0030116), alpha3 (HLA-G4; skipping of exon 4, HsaEX6046105), or both the alpha2 and alpha3 domains (HLA-G3).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types