RnoEX6061306 @ rn6
Exon Skipping
Gene
ENSRNOG00000049590 | RT1-M2
Description
RT1 class Ib, locus M2 [Source:RGD Symbol;Acc:3493]
Coordinates
chr20:1337503-1339228:-
Coord C1 exon
chr20:1338959-1339228
Coord A exon
chr20:1338463-1338738
Coord C2 exon
chr20:1337503-1337778
Length
276 bp
Sequences
Splice sites
3' ss Seq
AACTTGTGGGACGGAACCAGGTT
3' ss Score
-0.51
5' ss Seq
CAGGTACAA
5' ss Score
7.09
Exon sequences
Seq C1 exon
GCTCCCACTCCTTGCGGTACTTTGACATCGCAGTGTCAAGGCCTGGCCTAGAGGAGACCCACTACATGACTCTTGGCTACGTGGATGACACAGAGTTTGTGCATTTCAACAATGAGGCAGTGAATCCGAGGTTTGAGCCCCGGGTGCCCTGGATGGAACAGATGGGACAGAAGTACTGGGATGACCAGACACGAATTGCCAAAGTCGCGGAACAGCAGATTAGAGTGTACTTTCAGAAACTGCGAGGCTACTACAACCAGAGCCAGAACA
Seq A exon
GTTCTCACACCATCCAAAGGATGACTGGCTGCTACATTGGACCAGACGGACACCTACTTCACGCATACCGTCAGTTTGGATATGATGGACAAGATTACCTCACTTTGAATGAAGATCTGAGCACCTGGACTGCGGCAGATTCGGCAGCTGAGATCACCCATAGAGAGTGGAAGGCAACTAATGAAGCTGAGTTCTGGAGGGTCTACTTGGAGGGCCCTTGCATGGTGTGGCTCTTTAAATACCTGACAGTGGGAAATGAGACTCTACTGCGCGCAG
Seq C2 exon
ATCCCCCCAAAGTATATGTGACCCATCACCCCAGACCTGAAGGTGATATCACCCTGAGGTGCTGGGCCCTGGGCTTCTACCCTTCTGACATCACCCTGATCTGGCAAAGAGATGGAGAGGACCTGACCCAGGACATGGACGTAATTGAGACCAGACCTGCAGGGGATGGAACCTTCCAGAAGTGGGCAGCTGTGGTGGTGCCTTCGGGGAAGGAGCAGAATTATACATGCTATGTAGAACATGAGGGGCTGCCTGAACCCCTCACCCTGAGATGGA
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000049590-'1-2,'1-1,2-2=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.011 A=0.011 C2=0.043
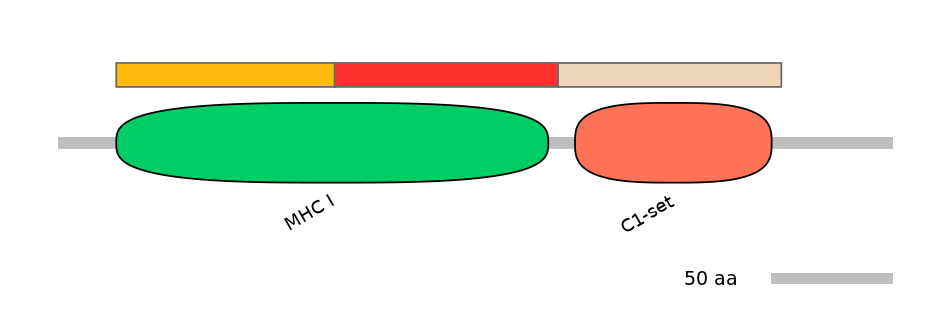
Domain overlap (PFAM):
C1:
PF0012913=MHC_I=PU(50.3=98.9)
A:
PF0012913=MHC_I=PD(49.2=94.6)
C2:
PF0765410=C1-set=WD(100=88.2)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGCGAGGCTACTACAACCAGA
R:
ATCTCAGGGTGAGGGGTTCAG
Band lengths:
303-579
Functional annotations
There are 2 annotated functions for this event
PMID: 11290782
Four membrane-bound (HLA-G1 to -G4) and two soluble (HLA-G5, and -G6) proteins are generated by alternative splicing. Only HLA-G1 has been extensively studied in terms of both expression and function. The authors provide evidence here that HLA-G2, -G3, and -G4 truncated isoforms reach the cell surface of transfected cells, as endoglycosidase H-sensitive glycoproteins, after a 2-h chase period. Moreover, cytotoxicity experiments show that these transfected cells are protected from the lytic activity of both innate (NK cells) and acquired (CTL) effectors, although to slightly different levels. The full-length transcript (HLA-G1) encodes three extracellular domains (alpha1, alpha2, and alpha3), a transmembrane region, and a cytoplasmic tail. Three other spliced transcripts encode membrane-bound proteins that lack alpha2 (HLA-G2; skipping exon 3, HsaEX0030116), alpha3 (HLA-G4; skipping of exon 4, HsaEX6046105), or both the alpha2 and alpha3 domains (HLA-G3).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]