MmuEX0030353 @ mm9
Exon Skipping
Gene
ENSMUSG00000030739 | Myh14
Description
myosin, heavy polypeptide 14 [Source:MGI Symbol;Acc:MGI:1919210]
Coordinates
chr7:51889729-51893249:-
Coord C1 exon
chr7:51893135-51893249
Coord A exon
chr7:51890824-51890946
Coord C2 exon
chr7:51889729-51889916
Length
123 bp
Sequences
Splice sites
3' ss Seq
CCTCATCTGCCCATCCCCAGAAC
3' ss Score
8.37
5' ss Seq
CGGGTGAGG
5' ss Score
8.48
Exon sequences
Seq C1 exon
GTTGACTACAAAGCCAGTGAGTGGCTGATGAAGAACATGGACCCACTGAATGACAATGTGGCCGCCTTGCTTCACCAGAGCACGGATCGTCTCACAGCTGAGATCTGGAAGGATG
Seq A exon
AACAAGGGGGTCTCCAACAGTTCACTTTACTTGGCTCCTTCCCATCGCCGTCCCCTGGACCTGCAGGGAGACTCGGCTCTGGCGCTTCTCCTCCAGGGGTTGGGTCTCTCTGTGCACCCACGG
Seq C2 exon
TGGAGGGCATCGTGGGGCTGGAGCAAGTAAGCAGCCTTGGAGATGGCCCACCGGGAGGCCGCCCCCGCCGTGGAATGTTCCGGACTGTGGGGCAGCTCTACAAAGAATCCCTGAGCCGCCTCATGGCCACGCTCAGCAACACCAACCCTAGTTTTGTCCGCTGCATCGTTCCCAATCATGAGAAGAGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000030739_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.019 A=0.571 C2=0.258
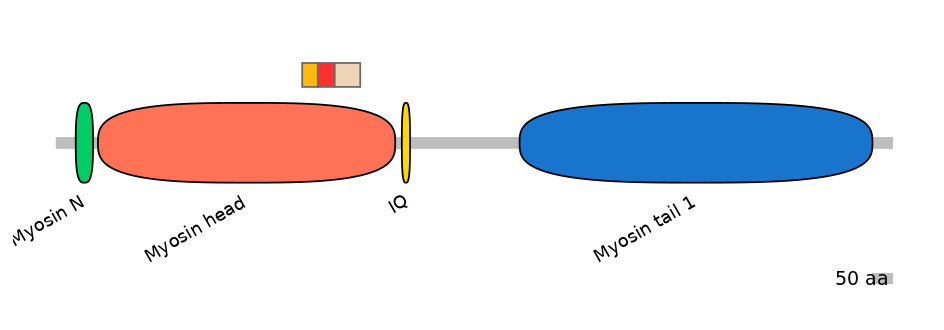
Domain overlap (PFAM):
C1:
PF0006316=Myosin_head=FE(5.5=100)
A:
PF0006316=Myosin_head=FE(5.7=100)
C2:
PF0006316=Myosin_head=FE(9.0=100)
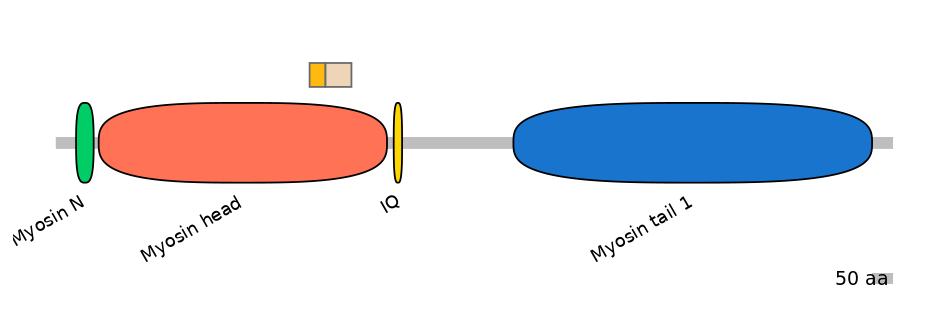
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACTGAATGACAATGTGGCCGC
R:
GAACGATGCAGCGGACAAAAC
Band lengths:
242-365
Functional annotations
There are 1 annotated functions for this event
PMID: 19240025
An isoform of non-muscle myosin II-C (NM II-C), called NM II-C2, is generated by inclusion of a casette exon, C2 (MmuEX0030353 in mouse, HsaEX0040957 in humans), encoding 41 amino acids in mice (33 amino acids in humans). The 41 amino acids are inserted into loop 2 of the NM II-C heavy chain within the actin binding region. Unlike most vertebrate non-muscle and smooth muscle myosin IIs, baculovirus-expressed mouse heavy meromyosin (HMM) II-C2 demonstrates no requirement for regulatory myosin light chain (MLC(20)) phosphorylation for maximum actin-activated MgATPase activity or maximum in vitro motility as measured by the sliding actin filament assay. In contrast, noninserted HMM II-C0 and another alternatively spliced isoform HMM II-C1, which contains 8 amino acids inserted into loop 1, are dependent on MLC(20) phosphorylation for both actin-activated MgATPase activity and in vitro motility. HMM II-C1C2, which contains both the C1 and C2 inserts, does not require MLC(20) phosphorylation for full activity similar to HMM II-C2. These constitutively active C2-inserted isoforms of NM II-C are expressed only in neuronal tissue. This is in contrast to NM II-C1 and NM II-C0, both of which are ubiquitously expressed. Full-length NM II-C2-GFP expressed in COS-7 cells localizes to filaments in interphase cells and to the cytokinetic ring in dividing cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: