RnoALTA0034041-2/3 @ rn6
Alternative 3'ss
Gene
ENSRNOG00000002364 | Rnf112
Description
ring finger protein 112 [Source:RGD Symbol;Acc:3986]
Coordinates
chr10:47723054-47723518:-
Coord C1 exon
chr10:47723233-47723518
Coord A exon
chr10:47723087-47723089
Coord C2 exon
chr10:47723054-47723086
Length
3 bp
Sequences
Splice sites
5' ss Seq
CAGGTGAGG
5' ss Score
10.07
3' ss Seq
CCCTTCTCTGTCCTGCCCAGGAG
3' ss Score
9.52
Exon sequences
Seq C1 exon
GTCCCATGCATCATTCCCCAAGCTGGAGCTGGGCCTGGGACAGCGTCCCTCCCCACCCCGGGAGTCGCCTACCTGCTCCATCTGTCTGGAAAGGCTTCGAGAGCCTATCTCACTGGACTGTGGCCACGACTTCTGCATCCGATGCTTCAGCACACATCGCATCCCAGGCTGTGAGCTGCCATGCTGTCCTGAATGCCGGAAAATCTGTAAGCAAAAGAAGGGCCTTCGCAGTCTAGGGGAGAGGATGAAGCTCCTACCTCAGCGGCCGCTGCCCCCTGCACTGCAG
Seq A exon
GAG
Seq C2 exon
ACCTGTGCTGTGAGAGCGGAGCGTCTGCTGTTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000002364-11-11,11-10,11-9-2/3
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.089 A=NA C2=0.000
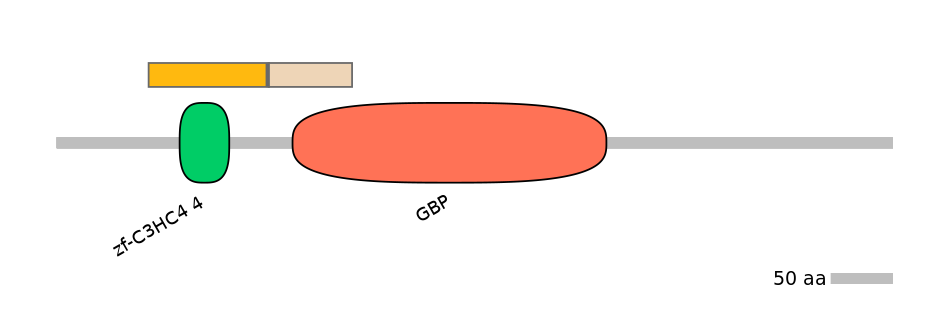
Domain overlap (PFAM):
C1:
PF152271=zf-C3HC4_4=WD(100=42.7)
A:
NA
C2:
PF0226314=GBP=PU(18.9=69.6)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AAAAGAAGGGCCTTCGCAGTC
R:
AGACGCTCCGCTCTCACAG
Band lengths:
100-103
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]