RnoEX0042387 @ rn6
Exon Skipping
Gene
ENSRNOG00000008313 | Hoxb3
Description
homeo box B3 [Source:RGD Symbol;Acc:1310780]
Coordinates
chr10:84190087-84194566:+
Coord C1 exon
chr10:84190087-84190174
Coord A exon
chr10:84193576-84193639
Coord C2 exon
chr10:84194466-84194566
Length
64 bp
Sequences
Splice sites
3' ss Seq
ACTCCACCCTCACCAAACAGATA
3' ss Score
8.23
5' ss Seq
CAGGTACCA
5' ss Score
7.88
Exon sequences
Seq C1 exon
GAGGGGAGATTTCTCGCCTGCCGCTCGCGCTGGGGCTCGATGTGAATATATATTATGTCTGCCTGTTCTCCCCTCGTCGGTGGCTAAG
Seq A exon
ATATTCCCCTGGATGAAAGAGTCAAGGCAAACGTCCAAGCTGAAAAACAGCTCCCCGGGCACAG
Seq C2 exon
CAGAAGGTTGTGGTGGCGGCGGCGGCGGCGGTGGCGGCGGCGGTAGTAGCAGCGGCGGGGGCGGCAGCGGCGGCGGGGGAGGGGACAAGAGCCCCCCGGGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000008313-'5-6,'5-5,6-6
Average complexity
C2*
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.840 C2=NA
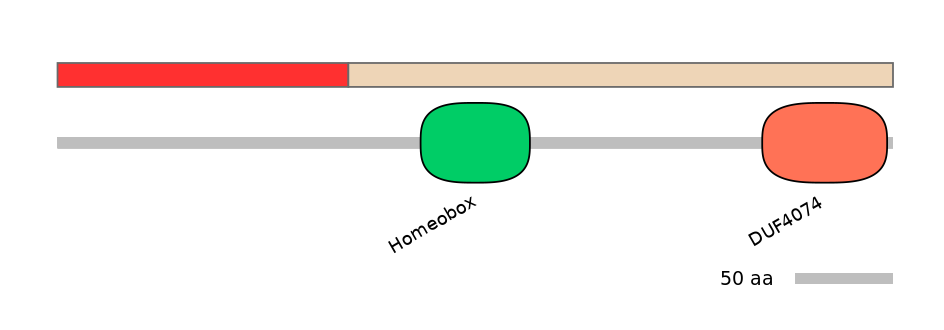
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
NA
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TATTATGTCTGCCTGTTCTCCCCT
R:
GGGCTCTTGTCCCCTCCC
Band lengths:
132-196
Functional annotations
There are 1 annotated functions for this event
PMID: 8600458
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: not specified. ELM ID: ELMI000757; ELM sequence: FPWM; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]