RnoEX0056910 @ rn6
Exon Skipping
Gene
ENSRNOG00000049695 | Myh1
Description
myosin heavy chain 1 [Source:RGD Symbol;Acc:735061]
Coordinates
chr10:53749667-53751344:+
Coord C1 exon
chr10:53749667-53749979
Coord A exon
chr10:53750097-53750171
Coord C2 exon
chr10:53751257-53751344
Length
75 bp
Sequences
Splice sites
3' ss Seq
CTTTCCTTGCTTCCCCAGAGTCT
3' ss Score
-3.52
5' ss Seq
AGGGTACAA
5' ss Score
1.91
Exon sequences
Seq C1 exon
CCAATGGGCATCTTCTCCATCCTGGAAGAGGAGTGCATGTTCCCCAAAGCGACAGACACCTCCTTCAAGAACAAGCTGTATGAACAGCATCTTGGAAAGTCCAACAACTTCCAGAAGCCCAAACCCGCCAAAGGCAAGGTTGAGGCCCACTTCTCTCTGGTGCACTATGCCGGCACCGTTGACTACAACATCGCTGGCTGGCTGGACAAGAACAAGGACCCCCTGAATGAGACCGTGGTGGGGCTGTACCAGAAGTCTTCAATGAAAACTCTGGCTTACCTCTTCTCTGGGGCAGCAGCTGCTGCTGAAGCAG
Seq A exon
TCTGGTGGCGGTGGTGGAAAGAAAGGGGCAAAGAAGAAGGGTTCTTCCTTCCAGACTGTGTCAGCTCTCTTCAGG
Seq C2 exon
GAGAACCTGAATAAGCTGATGACCAATCTGAGGAGCACTCACCCCCACTTTGTCCGGTGCATCATCCCCAATGAGACTAAGACGCCTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000049695-'62-68,'62-67,63-68
Average complexity
S
Mappability confidence:
75%=75=75%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.029 A=0.404 C2=0.000
Domain overlap (PFAM):
C1:
PF0006316=Myosin_head=FE(15.1=100)
A:
PF0006316=Myosin_head=FE(3.6=100)
C2:
PF0006316=Myosin_head=FE(4.2=100)
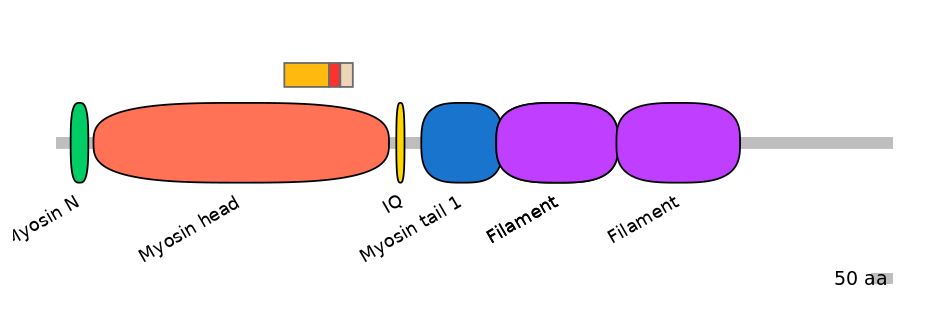
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACAAGAACAAGGACCCCCTGA
R:
GGATGATGCACCGGACAAAGT
Band lengths:
175-250
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]