RnoEX0077193 @ rn6
Exon Skipping
Gene
ENSRNOG00000015701 | Rreb1
Description
ras responsive element binding protein 1 [Source:RGD Symbol;Acc:1310904]
Coordinates
chr17:27543418-27558973:-
Coord C1 exon
chr17:27556057-27558973
Coord A exon
chr17:27550106-27550267
Coord C2 exon
chr17:27543418-27544224
Length
162 bp
Sequences
Splice sites
3' ss Seq
ATTTTTTTTTCCTGCTTCAGGTC
3' ss Score
11.01
5' ss Seq
CAGGTAGAG
5' ss Score
7.1
Exon sequences
Seq C1 exon
CACAGACAGCCCAGGAGGCTCCATCCCCAAGGCCTCCCCTAGCCCCACAGACATCCCAAGCTCGAGAGAACCCACTGACCCAGCCCCTACAGCCAGCAGCCCCGAGGAAGCCTCGCCTCCTGAACAAGGGCCGGCTGCCTCATCCAGGAAGAGGGGCAGGAAAAGGGGAATGAGAAACCGACCCCTCCCCAACAGCAGTGCTGTGGACCTGGACTCCAGTGGGGAGTTTGCCAGCATCGAGAAGATGCTGGCCACCACAGACACCAACAAGTTCAGTCCGTTCCTGCAGACGGCAGAGGACACTCAGGAAGAGGTGGCTGGAACCCCTGCAGACCACCGCGGGCCCACTGAGGAGGAGCAAGGCAGCTCTGCGGAGGACAGGCTGCTGAGAGCAAAGCGGAACTCCTATGCCAACTGCCTGCAGAAGATCAACTGTCCCCACTGTCCCCGGGTCTTCCCTTGGGCCAGCTCCCTGCAGAGGCACATGCTTACACACACTG
Seq A exon
GTCAGAAACCCTTCCCCTGTCAAAAATGCGATGCCTTCTTTTCTACCAAATCTAACTGTGAACGTCACCAGTTGCGCAAACACGGAGTTACCACCTGCTCCCTGAGAAGAAACGGGCTTATTCCCCCAAAAGAGGGTGATGTTGGATCCCATGATAGCACAG
Seq C2 exon
ACAGCCAGTCAGACACAGACACACTGACCACCCCAGGTGAGGTGCTGGATCTCACAGCACGGGACAAAGAGCAGCCAACATCAGAGGGAGCCAGTGAGCTCAGCCCAGCTACGCAGGACCTTACAGAAACAGAGGCCAAGGCAGAAGCAGCCCCACCTGAGGAGGAGGAGGAGGAAAAGGGGACGGAGGAGAACCCAGAACCAGAGTTAGAGTGCCGCGTGGAAGAGAGCTCTGGGGATGCAGACGCCCCGGAGGAAGACACAGCCAGCAACCAGAGCCTGGATCTTGACTTCGCCACCAAGATGATGGACTTCAAGCTGGCCGAGAGTGAGGCAGGTGCTGTGGACAGCCAAGGCCCGGCCCAGCAGGAGCCGAAGCATGCCTGCAATACCTGTGGGAAGAACTTCAAGTTCCTGAGCACCCTAAGTCGCCACAGGAAGGCGCACGGCTGCCAGGAGCCCAAGGAGGAGGAGGAGGCGCCTTCGCTGGAGAATGAGAGC
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000015701_MULTIEX1-2/2=1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
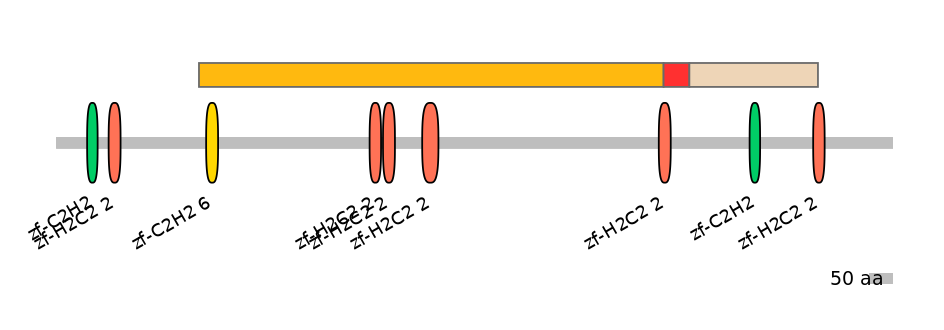
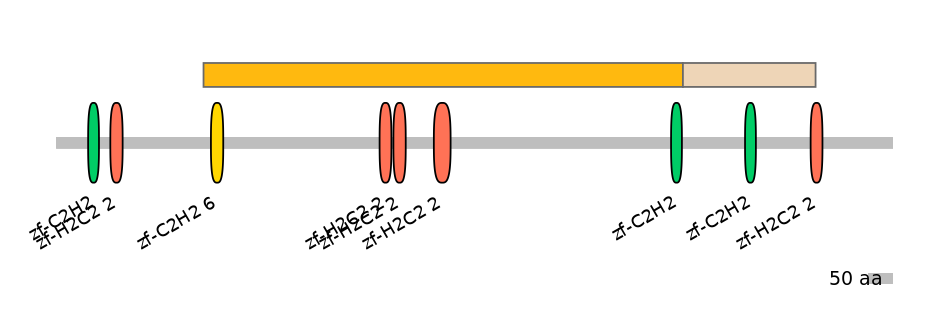
Features
Disorder rate (Iupred):
C1=0.356 A=0.455 C2=0.926
Domain overlap (PFAM):
C1:
PF139121=zf-C2H2_6=WD(100=2.7),PF134651=zf-H2C2_2=WD(100=2.6),PF134651=zf-H2C2_2=WD(100=2.7),PF134651=zf-H2C2_2=WD(100=3.6),PF134651=zf-H2C2_2=PU(38.5=1.0)
A:
PF134651=zf-H2C2_2=PD(57.7=27.3)
C2:
PF0009621=zf-C2H2=WD(100=8.5),PF134651=zf-H2C2_2=PU(40.0=3.7)
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGCAAAGCGGAACTCCTATGC
R:
CTTCTGCCTTGGCCTCTGTTT
Band lengths:
257-419
Functional annotations
There are 1 annotated functions for this event
PMID: 21703425
In vitro proliferation studies using the UMUC-3 bladder cancer cell line after selective isoform-specific knockdown of expression indicate that RREB1alpha (all included) is not necessary for proliferation, but that RREB1beta (skipping exon 8, HsaEX0055741) may be required.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]