RnoEX6042599 @ rn6
Exon Skipping
Gene
ENSRNOG00000003840 | Slit2
Description
slit guidance ligand 2 [Source:RGD Symbol;Acc:69310]
Coordinates
chr14:66922362-66926931:-
Coord C1 exon
chr14:66926768-66926931
Coord A exon
chr14:66925883-66925906
Coord C2 exon
chr14:66922362-66922512
Length
24 bp
Sequences
Splice sites
3' ss Seq
CTCTCTCTTTTTTTATTTAGCTA
3' ss Score
8.68
5' ss Seq
CAGGTAGGT
5' ss Score
10.28
Exon sequences
Seq C1 exon
GCATTTGGCCCAGAATCCTTTCATTTGTGACTGCCATCTCAAGTGGCTAGCGGATTATCTCCACACCAACCCAATTGAGACCAGCGGTGCCCGTTGCACCAGTCCCCGCCGCCTGGCTAACAAAAGAATTGGACAGATCAAAAGCAAGAAATTCCGTTGTTCAG
Seq A exon
CTAAAGAGCAGTATTTCATTCCAG
Seq C2 exon
GTACAGAAGATTATCGATCAAAATTAAGTGGAGACTGCTTTGCAGACTTGGCTTGTCCTGAAAAATGTCGCTGTGAAGGGACCACAGTAGACTGCTCCAATCAAAAACTCAACAAAATCCCAGACCATATTCCCCAGTACACAGCAGAGCT
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000003840-'18-22,'18-20,19-22=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF138551=LRR_8=PD(11.5=12.5)
A:
NO
C2:
PF0146213=LRRNT=WD(100=54.9),PF138551=LRR_8=PU(5.0=5.9)
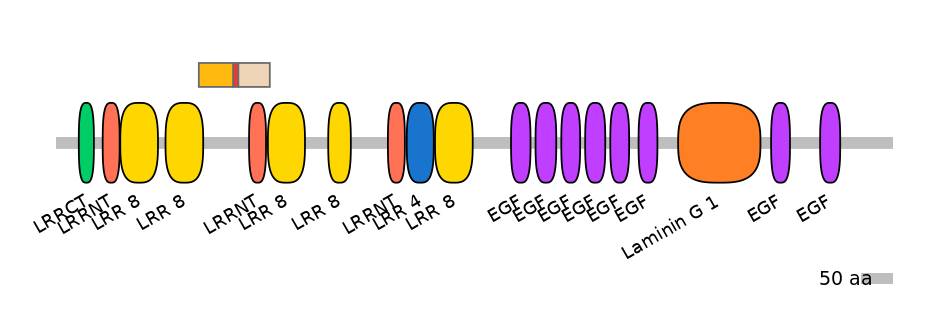
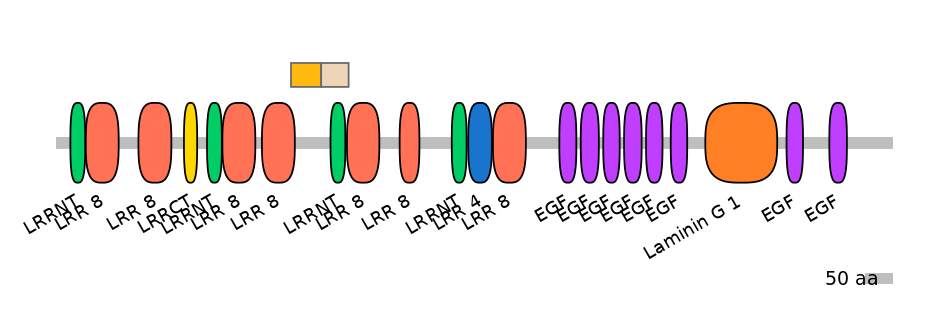
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCGCCTGGCTAACAAAAGAAT
R:
CAGGACAAGCCAAGTCTGCAA
Band lengths:
116-140
Functional annotations
There are 3 annotated functions for this event
PMID: 21264840
Exon 15 (HsaEX0060159) encodes a region of the Slit2 protein near the binding domain of the Robo1 receptor. The exclusion of exon 15 inhibits lung cancer cell growth in cell lines and mouse xenografts. Both isoforms of Slit2, with and without exon 15, suppress cancer invasiveness. In vivo, the inclusion of this exon may be an important event in cancer progression and invasion.
PMID: 26021305
The aim of this study was to elucidate the differential roles of these exon 15 splicing variants in angiogenesis. These results revealed that both Slit2-WT and Slit2-deltaE15 inhibit motility of human umbilical vein endothelial cells (HUVECs). The conditioned medium (CM) collected from CL1-5/VC or CL1-5/Slit2-WT lung adenocarcinoma cells blocked HUVEC tube formation and angiogenesis on chorioallantoic membrane (CAM) assay when compared with untreated HUVECs and CAM, respectively. However, CM of CL1-5/Slit2-deltaE15 restored the quality of tubes and the size of vessels. Although both Slit2-WT and Slit2-deltaE15 inhibited permeability induced by CM of cancer cells, Slit2-deltaE15 exhibited stronger effect. These results suggested that Slit2-deltaE15 plays important roles in normalization of blood vessels by enhancing tube quality and tightening endothelial cells, while Slit2-WT only enhances tightening of endothelial cells. It appears that Robo4 is responsible for Slit2 isoform-mediated inhibition of permeability, while neither Robo1 nor Robo4 is required for Slit2-deltaE15-enhanced tube quality.
PMID: 30717252
The results of this study suggested that the upregulation of Slit2-WT, but not Slit2-deltaE15, in a cancer microenvironment is an important factor in suppressing immunity while not interfering with cancer growth. These conclusions are based on indirect assays: testing how the ratio inclusion/skipping changes in different experimental conditions.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]