BtaINT0058887 @ bosTau6
Intron Retention
Gene
ENSBTAG00000018872 | F12
Description
coagulation factor XII (Hageman factor) [Source:HGNC Symbol;Acc:HGNC:3530]
Coordinates
chr7:40259878-40260407:-
Coord C1 exon
chr7:40260196-40260407
Coord A exon
chr7:40260098-40260195
Coord C2 exon
chr7:40259878-40260097
Length
98 bp
Sequences
Splice sites
5' ss Seq
CAGGTAGTT
5' ss Score
6.3
3' ss Seq
AGCCTGGGCTCCTCCCACAGGCT
3' ss Score
7.56
Exon sequences
Seq C1 exon
CGCCTCCACCCCACCTCGTGGTTACAGGAACCCCGACAACGACACCCGCCCTTGGTGCTTCATTTGGAAAGGCGACCGACTGAGCTGGAATTACTGCCGCCTGGCACCGTGCCAGGCCGCAGCTGGGCACGAGCACTTCCCCTTGCCCTCGCCATCGGCTTTGCAGAAACCTGAGTCCACGACCCAGACCCCGCTTCCATCCCTGACTTCAG
Seq A exon
GTAGTTGGGAAGGGGCGAGCGTAGAGGGCGTAGGACGAGCTATATTCCAACCGATCGACAGTGGGTCTCCAGGTCCTCAGCCTGGGCTCCTCCCACAG
Seq C2 exon
GCTGGTGCTCGCCGACTCCTCTGGCCAGCGGAGGCCCCGGGGGCTGTGGCCAGCGGCTCCGCAAATGGCTGTCCTCGCTGAACCGCGTCGTCGGAGGACTGGTGGCGCTCCCCGGGGCGCACCCCTACATCGCCGCGCTGTACTGGGACCAACATTTCTGCGCCGGCAGCCTCATCGCTCCCTGTTGGGTGCTGACTGCGGCTCACTGTCTGCAGAACCG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000018872:ENSBTAT00000025122:9
Average complexity
IR
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.222 A=NA C2=0.000
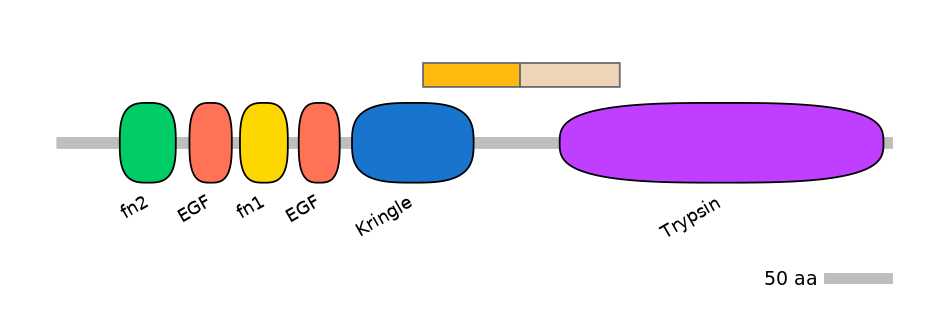
Domain overlap (PFAM):
C1:
PF0005113=Kringle=PD(41.1=51.4)
A:
NA
C2:
PF0008921=Trypsin=PU(18.5=59.5)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGCTTCCATCCCTGACTTCAG
R:
CGCAGAAATGTTGGTCCCAGT
Band lengths:
183-281
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]