DmeEX0003484 @ dm6
Exon Skipping
Gene
FBgn0020503 | CLIP-190
Description
The gene Cytoplasmic linker protein 190 is referred to in FlyBase by the symbol DmelCLIP-190 (CG5020, FBgn0020503). It is a protein_coding_gene from Dmel. It has 16 annotated transcripts and 16 polypeptides (15 unique). Gene sequence location is 2L:17384700..17409698. Its molecular function is described by: microtubule binding; myosin VI heavy chain binding; actin binding; protein binding; microtubule plus-end binding. It is involved in the biological process described with: nuclear migration; cytoplasmic microtubule organization; cellularization; establishment of mitotic spindle orientation. 21 alleles are reported. No phenotypic data is available. The phenotypic classes of alleles include: viable; fertile. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of low expression. Peak expression observed within 18-24 hour embryonic stages, at stages throughout the pupal period, in adult male stages.
Coordinates
chr2L:17385561-17400551:+
Coord C1 exon
chr2L:17385561-17385895
Coord A exon
chr2L:17394430-17394492
Coord C2 exon
chr2L:17400396-17400551
Length
63 bp
Sequences
Splice sites
3' ss Seq
GTTTTTCATTTTATTTTTAGAAG
3' ss Score
7.51
5' ss Seq
CAGGTAAAT
5' ss Score
8.76
Exon sequences
Seq C1 exon
TTTTTTAAATCGTTTACTTTTAATACGCGTGTGTCCTAATAAGTGAAGATTATTAAGAATAAAGTAAAAAATGAGTGATGATACGAGCGCTTCGGGCGGGACAAGTGCCCCTTTTCCATCGCCAGTTACTGCGGACCCGGAACCAGGAGCCACTGCCTCCAAATTACCGGGTCCCATCAGATCCAATATTCCCACGCCCGCTACTTCTGGTACCGGAATCCCGCAGCCCAGCAAAATGAAGGCACCATCTAGTTTTGGATCGACGGGTTCTGTTTCCAAAATCGGAAGACCGTGCTGCAATCACACAACTCCCAAATCCGGTCCACCACCAAGAG
Seq A exon
AAGCCACCAGCATGAGTCGTGAAAGCGATGACAATTTAAGTTCGATCAATTCGGCTTATACAG
Seq C2 exon
ATAACAGCAGCGCCGTGCTGACAGCAAACACAGAGCAGTTCATCATTGGACAGCGGGTTTGGCTGGGTGGCACTCGTCCCGGACAGATTGCCTTCATTGGAGACACACACTTTGCTGCTGGCGAATGGGCGGGTGTTGTCCTTGACGAGCCTAATG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0020503-'2-18,'2-11,5-18
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.972 A=0.621 C2=0.077
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0130220=CAP_GLY=PU(56.1=69.8)
Main Inclusion Isoform:
FBpp0080529
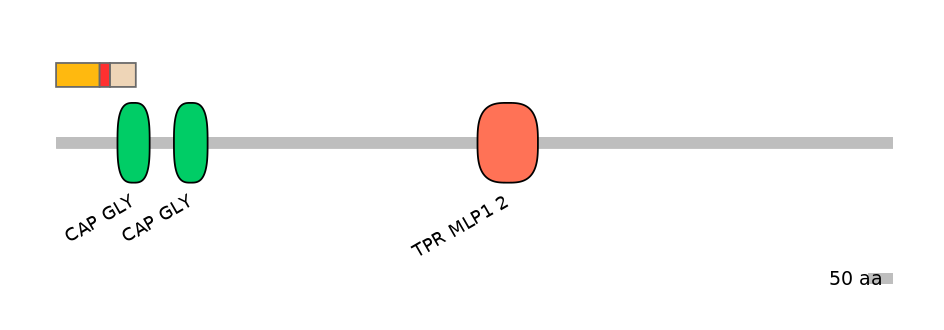
Main Skipping Isoform:
FBpp0290655
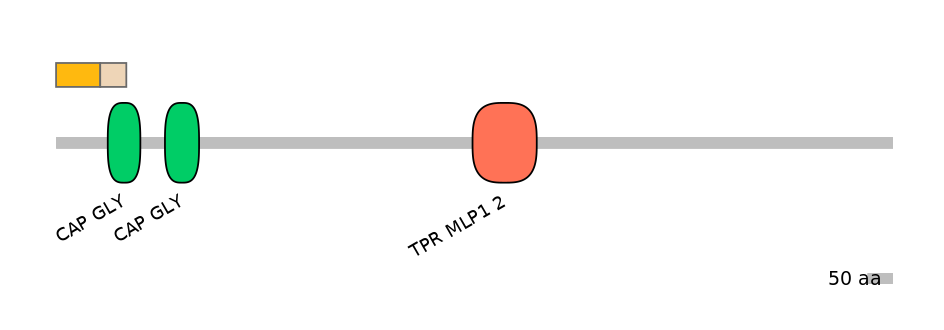
Other Inclusion Isoforms:
FBpp0080528, FBpp0080530, FBpp0288701, FBpp0288702, FBpp0290656, FBpp0290657, FBpp0304724, FBpp0304725, FBpp0304726, FBpp0304727, FBpp0304728, FBpp0308841
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGATCGACGGGTTCTGTTTCC
R:
ACCCGCTGTCCAATGATGAAC
Band lengths:
137-200
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)