DmeEX0004099 @ dm6
Exon Skipping
Gene
FBgn0033159 | Dscam1
Description
The gene Down syndrome cell adhesion molecule 1 is referred to in FlyBase by the symbol DmelDscam1 (CG17800, FBgn0033159). It is a protein_coding_gene from Dmel. It has 76 annotated transcripts and 76 polypeptides (75 unique). Gene sequence location is 2R:7317924..7381899. Its molecular function is described by: identical protein binding; antigen binding; protein homodimerization activity; axon guidance receptor activity; cell-cell adhesion mediator activity. It is involved in the biological process described with 16 unique terms, many of which group under: central nervous system development; response to other organism; cell-cell adhesion via plasma-membrane adhesion molecules; nervous system process; developmental growth. 159 alleles are reported. The phenotypes of these alleles manifest in: cell projection; plasma membrane bounded cell projection; neuron projection; mushroom body beta-lobe; larval neuromere. The phenotypic classes of alleles include: increased mortality; cell migration defective; phenotype; behavior defective. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of very low expression. Peak expression observed within 00-06 hour embryonic stages.
Coordinates
chr2R:7332261-7348010:-
Coord C1 exon
chr2R:7347849-7348010
Coord A exon
chr2R:7347392-7347697
Coord C2 exon
chr2R:7332261-7332380
Length
306 bp
Sequences
Splice sites
3' ss Seq
TACCCAAACCTCTCGCTCAGTTC
3' ss Score
4.11
5' ss Seq
ACGGTTCGC
5' ss Score
3.42
Exon sequences
Seq C1 exon
ATAACCGTGCCCTGCCCATCAACCGCAAACAGAAGGTCTTCCCCAACGGAACGCTGATTATCGAGAATGTGGAACGGAACTCAGACCAAGCGACCTACACTTGCGTTGCCAAGAATCAGGAAGGATACTCTGCTCGAGGATCTCTGGAAGTGCAAGTCATGG
Seq A exon
TTCCACCGCAGGTCGTACCCTTTGATTTCGGTGAGGAAACCATCAACATGAATGACATGGTCTCGGCCACGTGCACAGTGAACAAGGGCGACACTCCCCTGGAGCTGTACTGGACAACGGCTCCGGATCCCACGACGGGAGTGGGACGCCTGATGTCCAACGATGGCATTCTAATCACAAAGACGACGCAGCGCATCAGCATGCTGAGCATAGAGTCCGTGCATGCTCGCCATCGGGCGAACTACACGTGTGTGGCCAGGAATGCGGCCGGGGTCATCTACCACACGGCAGAGCTGCGCGTTAACG
Seq C2 exon
TACCTCCCAGATGGATCCTCGAGCCCACCGATAAGGCCTTCGCCCAGGGATCCGATGCCAAGGTTGAATGCAAGGCTGATGGCTTCCCCAAACCCCAAGTTACATGGAAGAAAGCAGTTG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0033159_MULTIEX1-1/49=C1-34
Average complexity
ME(1-5[100=84];1-6[100=91];1-8[100=84];1-9[100=93];1-10[100=85];1-13[100=89];1-20[100=82];1-22[100=87];1-28[100=82];1-30[100=94];1-31[100=92])
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.007 A=0.068 C2=0.178
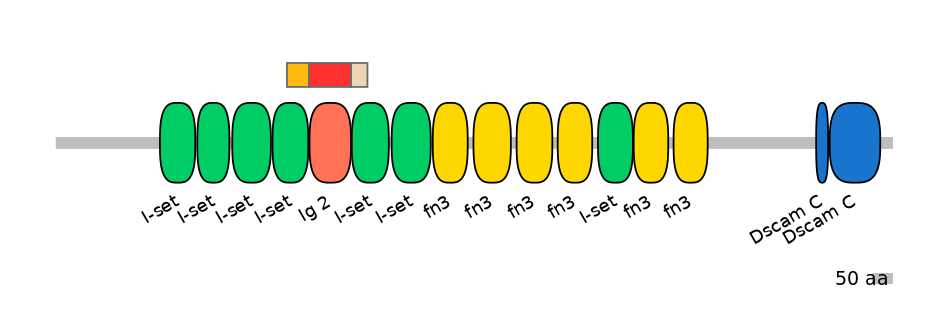
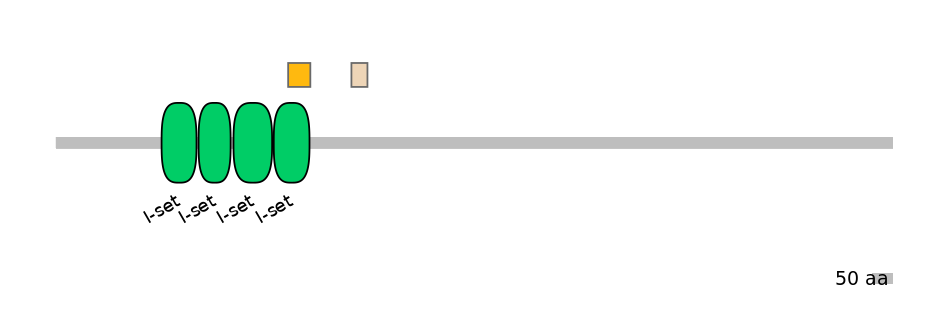
Domain overlap (PFAM):
C1:
PF0767911=I-set=PD(59.1=94.5)
A:
PF138951=Ig_2=WD(100=99.0)
C2:
PF138951=Ig_2=PD(0.1=0.0),PF0767911=I-set=PU(41.8=92.7)
Main Inclusion Isoform:
FBpp0110354
Main Skipping Isoform:
FBpp0297683
Other Inclusion Isoforms:
FBpp0089327, FBpp0297697
Other Skipping Isoforms:
FBpp0088088, FBpp0088090, FBpp0088091, FBpp0110341, FBpp0110342, FBpp0110343, FBpp0110344, FBpp0110345, FBpp0110346, FBpp0110347, FBpp0110348, FBpp0110349, FBpp0110350, FBpp0110351, FBpp0110352, FBpp0110353, FBpp0110355, FBpp0110356, FBpp0110357, FBpp0110358, FBpp0110359, FBpp0110360, FBpp0110361, FBpp0110362, FBpp0110363, FBpp0110364, FBpp0110365, FBpp0110366, FBpp0110367, FBpp0110368, FBpp0110369, FBpp0110370, FBpp0110371, FBpp0110372, FBpp0110373, FBpp0110374, FBpp0110375, FBpp0110376, FBpp0110377, FBpp0110378, FBpp0110379, FBpp0110381, FBpp0110382, FBpp0110384, FBpp0110387, FBpp0110389, FBpp0110391, FBpp0110392, FBpp0110393, FBpp0110394, FBpp0110395, FBpp0297684, FBpp0297685, FBpp0297686, FBpp0297687, FBpp0297688, FBpp0297689, FBpp0297690, FBpp0297691, FBpp0297692, FBpp0297693, FBpp0297694, FBpp0297695, FBpp0297696, FBpp0297698, FBpp0297699, FBpp0297700, FBpp0297701, FBpp0297702, FBpp0297703, FBpp0306847
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATAACCGTGCCCTGCCCATC
R:
TGCTTTCTTCCATGTAACTTGGGG
Band lengths:
278-584
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)