DreEX0013139 @ danRer10
Exon Skipping
Gene
ENSDARG00000077582 | ank3b
Description
ankyrin 3b [Source:ZFIN;Acc:ZDB-GENE-060621-2]
Coordinates
chr12:7612954-7617008:-
Coord C1 exon
chr12:7616798-7617008
Coord A exon
chr12:7616332-7616418
Coord C2 exon
chr12:7612954-7613037
Length
87 bp
Sequences
Splice sites
3' ss Seq
ATGTGCTGACCCGACTCTAGCAG
3' ss Score
3.15
5' ss Seq
AAGGTACTA
5' ss Score
7.52
Exon sequences
Seq C1 exon
CCAAGAGCTGAAGTCCTCAATGCCTGGAGTGCAAGCTGAGAGGCAAAATGGAGAACATCCAGAGGTGTTGTCGCCAGACGCCAGTGAGTCTTCCAGCAAACCAAAGGGCAAAGCAAGGAAAGATTCTGAGCGAGAGGAAGCAAGCAGGGAGGTTCTGGAGGACAGAGGACCAAACAACAGGGAGGAGGGCATCTCCGAGCCTAAAGTCCAG
Seq A exon
CAGGAAGCTCTTAGAGAGAATGATTCCAGCACTGATGAGGAAGAGACTGTCACTACCAGGGTGTATCGGCGGAGAGTGATTCTGAAG
Seq C2 exon
GGTGATCAGGCCAAAAACATCCCTGGTGAGACTGTCACAGAAGAGCAGTACACTGACGAAGATGGAAACTTAGTCACCAGAAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000077582_CASSETTE1
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=0.966 C2=0.888
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Inclusion Isoform:
ENSDART00000088100fB1183
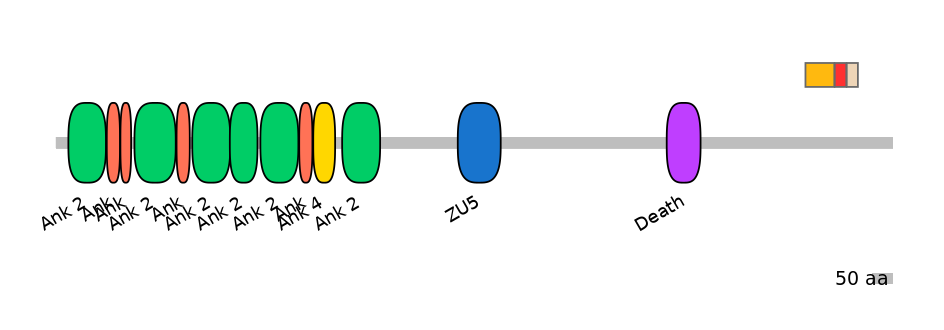
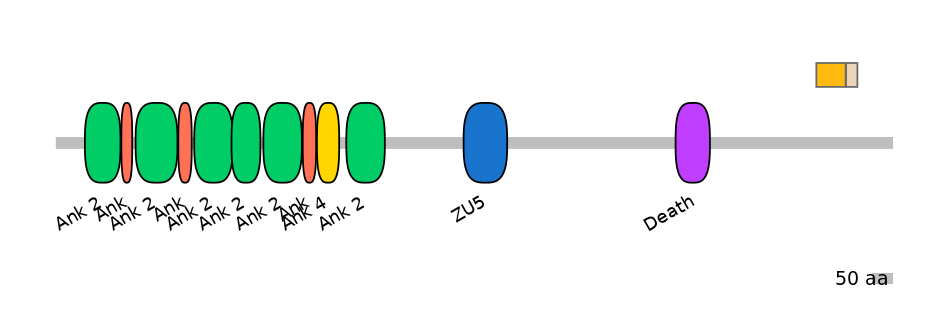
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCCAGTGAGTCTTCCAGCAAA
R:
TGACAGTCTCACCAGGGATGT
Band lengths:
169-256
Functional annotations
There are 1 annotated functions for this event
PMID: 21223964
Ankyrins-G coimmunoprecipitated with plectin and filamin C from skeletal muscle tissue extracts. Muscle-specific OTBD domain (coordinated inclusion of MmuEX1003810, MmuEX1003809, MmuEX0004908) is required for ankyrin-G binding to plectin and filamin C. Isoforms are differentially expressed during myogenesis.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]