DreEX0036199 @ danRer10
Exon Skipping
Gene
ENSDARG00000025728 | grin1b
Description
glutamate receptor, ionotropic, N-methyl D-aspartate 1b [Source:ZFIN;Acc:ZDB-GENE-051202-2]
Coordinates
chr5:29034079-29039335:-
Coord C1 exon
chr5:29039159-29039335
Coord A exon
chr5:29034948-29035010
Coord C2 exon
chr5:29034079-29034176
Length
63 bp
Sequences
Splice sites
3' ss Seq
CTGTGCACATTATTCATCAGAGT
3' ss Score
5.24
5' ss Seq
AAGGTATAT
5' ss Score
7.84
Exon sequences
Seq C1 exon
AGTATCCACCTCTCCTTCCTGCGCACGGTGCCTCCCTATTCCCACCAAGCCCAAGTGTGGTTTGACATGATGCGCGAGTTCCAGTGGAATCACATCATCCTGATTGTGAGCGACGACCACGAGGGCAGAGCGGCACAGAAAAGGCTGGAAACCCTTCTGGAGGAGAGGGAAACAAAG
Seq A exon
AGTAAAAACAGGAACTATGAAAACCTCGACCAACTGTCCTTTGACAACAAGCGAGGACCCAAG
Seq C2 exon
GCAGAGAAAGTCCTCCTGTTCAGCCAAGACACAAATCTGACGGCTCTGCTCCAGGAAGCCAAGGAGCTTGAGGCCCGAGTTATCATCCTGTCTGCAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000025728_CASSETTE3
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
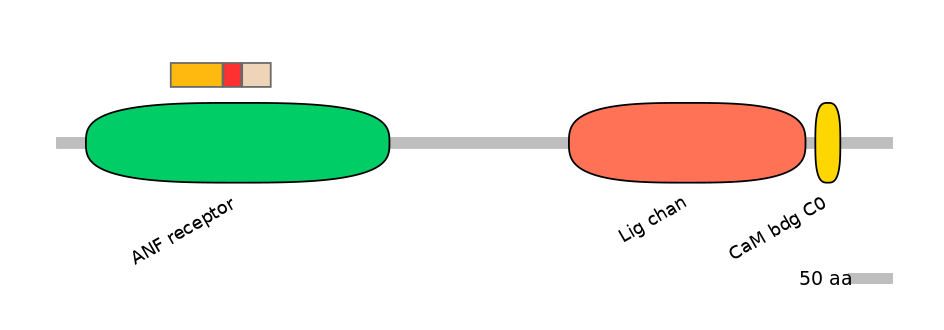
Features
Disorder rate (Iupred):
C1=0.144 A=0.381 C2=0.000
Domain overlap (PFAM):
C1:
PF0109423=ANF_receptor=FE(17.0=100)
A:
PF0109423=ANF_receptor=FE(5.9=100)
C2:
PF0109423=ANF_receptor=FE(10.0=100)
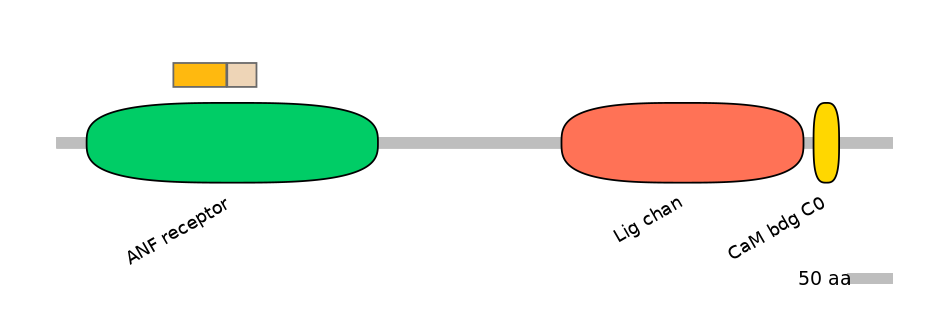
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCGAGTTCCAGTGGAATCACA
R:
TGTCTTGGCTGAACAGGAGGA
Band lengths:
135-198
Functional annotations
There are 10 annotated functions for this event
PMID: 8515865
In the presence of calcium, the reduction of total current was greatest in the NMDAR1-LL splice variant, and was significantly less in the NMDAR1-SS variant. The increased sensitivity of NMDAR1-LL may be attributed to a particularly sensitive slow current 'hump' which is more pronounced in NMDAR1-LL than in NMDAR1-SS. The ethanol sensitivity of these receptorsranged in order as NR1-1b (LL) > NR1-2b (LS) > NR1-1a (SL) > NR1-2a (SS), although this subunit dependence was lost whenrecordings were carried out in barium-containing media. LL = insertion of both HsaEX0028686 and HsaEX0028687. SS = skipping of both.
PMID: 10712457
This study performed patch-clamp recordings of L-glutamate responses from human embryonic kidney tumoral cells (HEK293) expressing NR1 subunit variants lacking exon 5 together with the NR2B subunit. These responses had deactivation components that lasted several seconds. The presence of exon 5 or spermine greatly accelerated deactivation of L-glutamate responses through alterations in desensitization. These effects were also observed at positive holding potentials and in the presence of physiological Mg(2+). Thus NR1 splicing and polyamines may have profound effects on the kinetics of NMDA receptor-mediated synaptic transmission.
PMID: 16573586
In this study, all 8 NR1 splice variants were individually coexpressed with each NR2 subunit in human embryonic kidney 293 (HEK293) cells and tested for inhibition by ethanol using patch-clamp electrophysiology. All 32 subunit combinations tested gave reproducible glutamate-activated currents and all receptors were inhibited to some degree by 100 mM ethanol. The sensitivity of individual receptors to ethanol was affected by the specific NR1 splice variant expressed with receptors containing the NR1-3 and NR1-4 subunits among the least inhibited by ethanol. The isoforms NR1-3 and NR1-4 could not be mapped (no info in the paper).
PMID: 22641781
GluN1-1b/GluN2D receptors, which contain the residues encoded by exon 5, deactivate with a dual exponential time course described by a _FAST of 410 ms and a _SLOW of 1100 ms. This time course is 3-fold more rapid than that for exon 5-lacking GluN1-1a/GluN2D, which deactivates with a _FAST of 1100 ms and a _SLOW of 3400 ms. Exon 5-containing NMDA receptors also have a two-fold higher open probability (0.037) than exon 5-lacking receptors (0.017). Furthermore, inclusion of exon 5-encoded residues within the GluN1-1b subunit decreases the potency for the endogenous agonist l-glutamate. Evaluation of receptor kinetics for NMDA receptors containing mutated GluN1-1b subunits and wild-type GluN2D identified residue Lys211 in GluN1-1b as a key determinant of exon 5 control of the deactivation time course and glutamate potency. Evaluation of a kinetic model of GluN1/GluN2D gating suggests that residues encoded by exon 5 influence several rate-limiting steps.
PMID: 7754371
Protons inhibit NMDA receptor function by 50 percent at pH 7.3 through interactions with the NR1 subunit, and both polyamines and NR1 exon 5 potentiate receptor function through relief of the tonic proton inhibition present at physiological pH. A single amino acid (lysine 211) was identified that mediates the effects of exon 5 in the rat brain. Electroneutral substitutions at this position restored pH sensitivity and, consequently, polyamine relief of tonic inhibition. This effect, together with the structural similarities between polyamines and the surface loop encoded by exon 5, suggest that exon 5 may act as a tethered pH-sensitive constitutive modulator of NMDA receptor function.
PMID: 10479681
When NR1 splice variants were expressed in fibroblasts, the amount of NR1 molecules expressed on the cell surface varied among forms with different C-terminal cytoplasmic domains. The splice forms with the longest C-terminal cytoplasmic tail (NR1-1a and NR1-1b) showed the lowest amount of cell surface expression, and the splice forms with the shortest C-terminal cytoplasmic tail (NR1-4a and NR1-4b) showed the highest cell surface expression. Cells transfected with 1b, 2b, 3b, or 4b demonstrate different levels of [Ca2+] increase following glutamate stimulation. N-cassette: MmuEX0021891??, C1: MmuEX0021891, C2-C2': MmuALTA0008176.
PMID: 29656875
It shows through cryoelectron microscopy (cryo-EM) that the presence of the exon 5 motif in GluN1 alters the local architecture of heterotetrameric GluN1-GluN2 NMDA receptors and creates contacts with the ligand-binding domains (LBDs) of the GluN1 and GluN2 subunits, which are absent in NMDA receptors lacking the exon 5 motif. The unique interactions established by the exon 5 motif are essential to the stability of the ATD/LBD and LBD/LBD interfaces that are critically involved in controlling proton sensitivity and deactivation.
PMID: 31570583
N-terminal splicing of GluN1 has important functions in the maturation of excitatory synapses. The inclusion of exon 5 of Grin1 is up-regulated in several brain regions such as the thalamus and neocortex. Deletion of Grin1 exon 5 disrupts the developmental remodeling of NMDARs in thalamic neurons and the effect is distinct from that of Grin2a (GluN2A) deletion. Deletion of Grin2a or exon 5 of Grin1 alone partially attenuates the shortening of NMDAR-mediated excitatory postsynaptic currents (NMDAR-EPSCs) during early life, whereas deletion of both Grin2a and exon 5 of Grin1 completely abolishes the developmental change in NMDAR-EPSC decay time. Deletion of exon 5 of Grin1 leads to an overproduction of excitatory synapses in layer 5 pyramidal neurons in the cortex and increases seizure susceptibility in adult mice.
PMID: 31875540
Here, the authors generate mice lacking from the GluN1 exon 5-encoded N1 cassette (GluN1a mice) or compulsorily expressing this exon (GluN1b mice). Despite no differences in basal synaptic transmission, long-term potentiation in the hippocampus is significantly enhanced in GluN1a mice compared with that in GluN1b mice. Furthermore, GluN1a mice learn more quickly and have significantly better spatial memory performance than do GluN1b mice. In addition, in human iPSC-derived neurons in autism spectrum disorder NMDARs show characteristics of N1-lacking GluN1. These findings indicate that alternative splicing of GluN1 is a mechanism for controlling physiological long-lasting synaptic potentiation, learning, and memory.
PMID: 34187890
The GluN1 subunit contains eight alternatively spliced isoforms produced by including or excluding the N1 and the C1, C2, or C2' polypeptide cassettes. Whether GluN1 alternative splicing affects nonionotropic signaling by NMDARs is a major outstanding question. Here, the authors discovered that glycine priming of recombinant NMDARs critically depends on GluN1 isoforms lacking the N1 cassette; glycine priming is blocked in splice variants containing N1. On the other hand, the C-terminal cassettes-C1, C2, or C2'-each permit glycine signaling. In wild-type mice, the authors found glycine-induced nonionotropic signaling at synaptic NMDARs in CA1 hippocampal pyramidal neurons. This nonionotropic signaling by glycine to synaptic NMDARs was prevented in mice the authors engineered, such that GluN1 obligatorily contained N1. The authors discovered in wild-type mice that, in contrast to pyramidal neurons, synaptic NMDARs in CA1 inhibitory interneurons were resistant to glycine priming. But the authors recapitulated glycine priming in inhibitory interneurons in mice engineered such that GluN1 obligatorily lacked the N1 cassette. Ex 5: MmuEX0021893, C-term cassettes: MmuALTA0008176 and MmuEX0021891.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]