DreEX6074500 @ danRer10
Exon Skipping
Gene
ENSDARG00000030364 | angpt1
Description
angiopoietin 1 [Source:ZFIN;Acc:ZDB-GENE-010817-1]
Coordinates
chr19:47231707-47264860:-
Coord C1 exon
chr19:47264759-47264860
Coord A exon
chr19:47257688-47257854
Coord C2 exon
chr19:47231707-47231837
Length
167 bp
Sequences
Splice sites
3' ss Seq
TCTCTCTCACTGTGTTGCAGGGT
3' ss Score
10.15
5' ss Seq
CAGGTACTG
5' ss Score
9.04
Exon sequences
Seq C1 exon
GTGTACTGTGTGATGGAGTCTGCTGGCGGAGGCTGGACGGTGATCCAGAAGCGTGAAGACGGCACTGTGGATTTCCAGAAAACATGGAAAGAATATAAGATG
Seq A exon
GGTTTTGGAAGCGTGTCGGGCGAACACTGGCTGGGGAACGAATTTGTCCACGTTCTGACCAATCAGAGGCAGCACGGCCTCCGCGTGGAGCTGAGTGACTGGGACGGACACCAGGCCTTCTCTCAGTACGACAGCTTCCACATCGACAGCGAGAAACAGAAATACAG
Seq C2 exon
GTTGTTTTTGAAGACCCACAGCGGGACGGCGGGACGGCAGAGCAGTTTGGCCATCCACGGAGCAGACTTCAGCACCAAAGACGTGGATAATGATAACTGCACATGCAAGTGTGCACTGATGCTCAGCGGCG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000030364-'5-8,'5-7,6-8=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
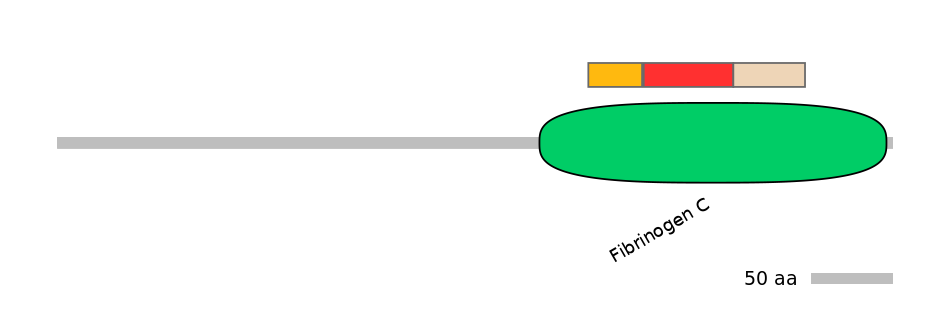
Domain overlap (PFAM):
C1:
PF0014713=Fibrinogen_C=FE(15.4=100)
A:
PF0014713=Fibrinogen_C=FE(25.7=100)
C2:
PF0014713=Fibrinogen_C=FE(20.6=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTGTGTGATGGAGTCTGCTGG
R:
GCATGTGCAGTTATCATTATCCACG
Band lengths:
203-370
Functional annotations
There are 1 annotated functions for this event
PMID: 10706866
Angiopoietin-1 (Ang-1) is required for developing vessels, and its absence leads to defects in vessel remodeling. Ang-1 has been identified as the ligand for the tyrosine kinase receptor Tie-2, which is expressed specifically on endothelial cells and early hematopoietic cells. In studying the role of Tie-2 and Ang-1 in megakaryocytopoiesis, 3 alternatively spliced species of Ang-1 mRNA (Ang-1.3 kb, Ang-0.9 kb, and Ang-0.7 kb) were identified in addition to the full-length Ang-1 (Ang-1.5 kb), in the megakaryocyte cell line CHRF by reverse transcription?polymerase chain reaction (RT-PCR), and then cloned and sequenced. The function of the 1.5-kb (contains exon encoding fibronogen C-terminal domain (HsaEX0004096)), 1.3-kb (lacks aforementioned exon), and 0.9-kb (contains the aforementioned exon) isoforms was examined. Recombinant proteins Ang-1.5 and 0.9 kb bind strongly to the recombinant Tie-2 receptor (Tie-2-Fc), whereas the 1.3-kb isoform does not. The Ang-1.3 kb isoform binds to the 1.5-kb isoform. Ang-1.5 kb, but not the 1.3-kb and 0.9-kb isoforms, induces tyrosine phosphorylation of Tie-2 in human umbilical vein endothelial cells. These data suggest that isoforms 1.3 kb and 0.9 kb could serve as dominant negative molecules for the full-length Ang-1. Of note, the only difference between the 1.5-kb and 1.3-kb isoforms is the presence/absence of the exon encoding the fibronogen C-terminal domain (HsaEX0004096).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]