HsaALTA1026631-2/2 @ hg38
Alternative 3'ss
Gene
ENSG00000134716 | CYP2J2
Description
cytochrome P450 family 2 subfamily J member 2 [Source:HGNC Symbol;Acc:HGNC:2634]
Coordinates
chr1:59907786-59909960:-
Coord C1 exon
chr1:59909784-59909960
Coord A exon
chr1:59907921-59907927
Coord C2 exon
chr1:59907786-59907920
Length
7 bp
Sequences
Splice sites
5' ss Seq
AAGGTGAGA
5' ss Score
8.68
3' ss Seq
TCAAACATTGTGTTTTCTAGCAC
3' ss Score
7.35
Exon sequences
Seq C1 exon
CTCTACAATGTCTTTCCATGGATAATGAAATTCCTGCCTGGACCCCACCAAACTCTCTTCAGCAACTGGAAAAAACTGAAATTGTTTGTTTCTCATATGATTGACAAACACAGAAAGGATTGGAATCCTGCAGAAACAAGAGACTTTATTGATGCTTACCTTAAAGAAATGTCAAAG
Seq A exon
CACACAG
Seq C2 exon
GCAATCCTACTTCAAGTTTCCATGAAGAAAACCTCATCTGCAGCACCCTGGACCTCTTCTTTGCCGGAACCGAGACAACTTCCACAACTCTGCGATGGGCTCTGCTTTATATGGCCCTCTACCCAGAAATCCAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000134716-12-11,12-10-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.000
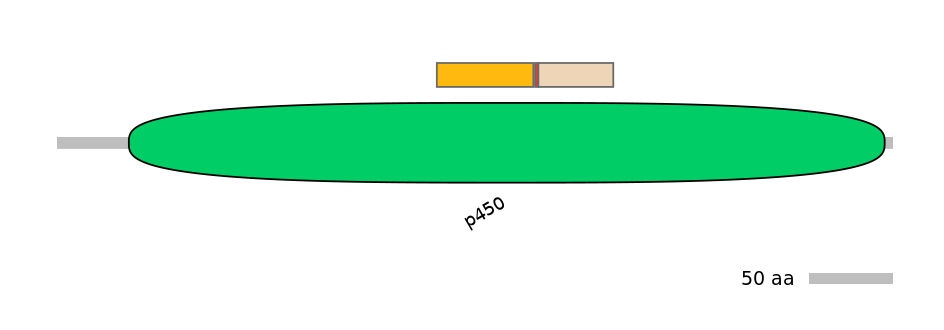
Domain overlap (PFAM):
C1:
PF0006717=p450=FE(12.7=100)
A:
NA
C2:
PF0006717=p450=FE(10.3=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGAATCCTGCAGAAACAAGAGAC
R:
GGGTGCTGCAGATGAGGTTTT
Band lengths:
105-112
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains