HsaALTD1026164-1/2 @ hg38
Alternative 5'ss
Gene
ENSG00000147140 | NONO
Description
non-POU domain containing, octamer-binding [Source:HGNC Symbol;Acc:HGNC:7871]
Coordinates
chrX:71297882-71298508:+
Coord C1 exon
chrX:71297882-71297938
Coord A exon
NA
Coord C2 exon
chrX:71298469-71298508
Length
0 bp
Sequences
Splice sites
5' ss Seq
GCGGTATAT
5' ss Score
5.01
3' ss Seq
GAGTATTTTTTGTTTTTCAGAGA
3' ss Score
8.49
Exon sequences
Seq C1 exon
CAAGAAGAAATGATGCGGCGACAGCAGGAAGGATTCAAGGGAACCTTCCCTGATGCG
Seq A exon
NA
Seq C2 exon
AGAGAGCAGGAGATTCGGATGGGTCAGATGGCTATGGGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000147140-37-35,38-35-1/2
Average complexity
Alt5
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=NA C2=1.000
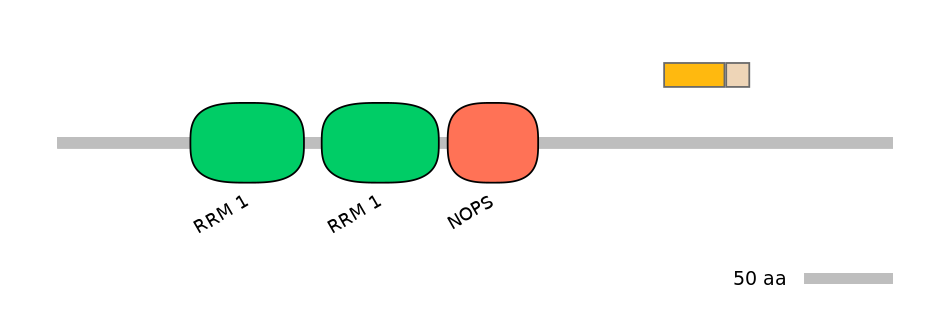
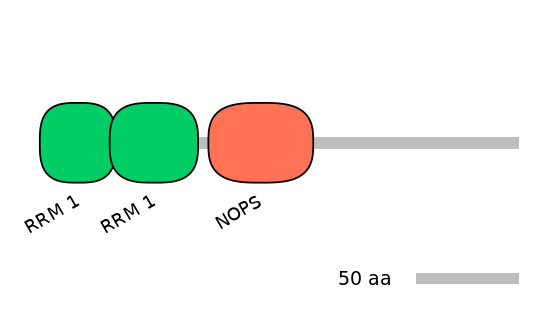
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
NO
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGAAGAAATGATGCGGCGACA
R:
CTCCCATAGCCATCTGACCCA
Band lengths:
95-235
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains