HsaEX0038125 @ hg19
Exon Skipping
Gene
ENSG00000076770 | MBNL3
Description
muscleblind-like splicing regulator 3 [Source:HGNC Symbol;Acc:20564]
Coordinates
chrX:131516206-131520839:-
Coord C1 exon
chrX:131520689-131520839
Coord A exon
chrX:131518691-131518726
Coord C2 exon
chrX:131516206-131516300
Length
36 bp
Sequences
Splice sites
3' ss Seq
CTTTTCTGCGTGGTGTACAGGGC
3' ss Score
7.59
5' ss Seq
TTGGTAGGT
5' ss Score
7.03
Exon sequences
Seq C1 exon
GCCCTGCAGCCTGGTACACTGCAACTGATACCAAAGAGATCAGCACTGGAAAAGCCCAATGGTGCCACCCCGGTCTTTAATCCCACTGTTTTCCACTGCCAACAGGCTCTGACTAACCTGCAGCTCCCACAGCCGGCATTTATCCCTGCAG
Seq A exon
GGCCAATACTGTGCATGGCACCCGCTTCAAATATTG
Seq C2 exon
TGCCCATGATGCACGGTGCTACACCTACCACTGTGTCTGCAGCAACAACACCTGCCACCAGCGTTCCGTTCGCTGCACCAACTACAGGCAATCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000076770_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
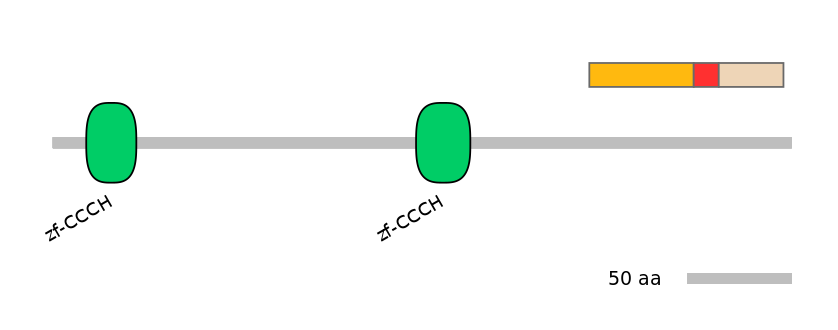
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.217 A=0.000 C2=0.686
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
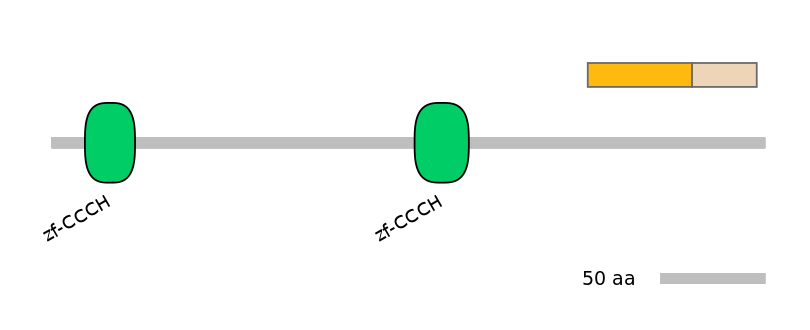
Other Inclusion Isoforms:
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATCCCACTGTTTTCCACTGCC
R:
TGTTGTTGCTGCAGACACAGT
Band lengths:
122-158
Functional annotations
There are 2 annotated functions for this event
PMID: 21454535
This event
Inferred from MBNL1 ortholog: The exon 7 region enhances MBNL1-MBNL1 dimerization properties
PMID: 27733504
This event
The presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (_2.7 and _4.0-times, respectively). There were marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (_1.3-times) and ex.95nt (_0.8-times). There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, exOFF events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)