HsaEX0066149 @ hg38
Exon Skipping
Gene
ENSG00000041982 | TNC
Description
tenascin C [Source:HGNC Symbol;Acc:HGNC:5318]
Coordinates
chr9:115059730-115064068:-
Coord C1 exon
chr9:115063796-115064068
Coord A exon
chr9:115062917-115063189
Coord C2 exon
chr9:115059730-115060002
Length
273 bp
Sequences
Splice sites
3' ss Seq
AACTCTGTGTCCTTATGCAGAGG
3' ss Score
7.58
5' ss Seq
CAGGTATAT
5' ss Score
7.88
Exon sequences
Seq C1 exon
GGGAAACTCCCAATTTGGGAGAGGTCGTGGTGGCCGAGGTGGGCTGGGATGCCCTCAAACTCAACTGGACTGCTCCAGAAGGGGCCTATGAGTACTTTTTCATTCAGGTGCAGGAGGCTGACACAGTAGAGGCAGCCCAGAACCTCACCGTCCCAGGAGGACTGAGGTCCACAGACCTGCCTGGGCTCAAAGCAGCCACTCATTATACCATCACCATCCGCGGGGTCACTCAGGACTTCAGCACAACCCCTCTCTCTGTTGAAGTCTTGACAG
Seq A exon
AGGAGGTTCCAGATATGGGAAACCTCACAGTGACCGAGGTTAGCTGGGATGCTCTCAGACTGAACTGGACCACGCCAGATGGAACCTATGACCAGTTTACTATTCAGGTCCAGGAGGCTGACCAGGTGGAAGAGGCTCACAATCTCACGGTTCCTGGCAGCCTGCGTTCCATGGAAATCCCAGGCCTCAGGGCTGGCACTCCTTACACAGTCACCCTGCACGGCGAGGTCAGGGGCCACAGCACTCGACCCCTTGCTGTAGAGGTCGTCACAG
Seq C2 exon
AGGATCTCCCACAGCTGGGAGATTTAGCCGTGTCTGAGGTTGGCTGGGATGGCCTCAGACTCAACTGGACCGCAGCTGACAATGCCTATGAGCACTTTGTCATTCAGGTGCAGGAGGTCAACAAAGTGGAGGCAGCCCAGAACCTCACGTTGCCTGGCAGCCTCAGGGCTGTGGACATCCCGGGCCTCGAGGCTGCCACGCCTTATAGAGTCTCCATCTATGGGGTGATCCGGGGCTATAGAACACCAGTACTCTCTGCTGAGGCCTCCACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000041982-'20-32,'20-27,22-32
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.084 A=0.207 C2=0.016
Domain overlap (PFAM):
C1:
PF0004116=fn3=WD(100=90.2)
A:
PF0004116=fn3=WD(100=88.0)
C2:
PF0004116=fn3=WD(100=87.0)
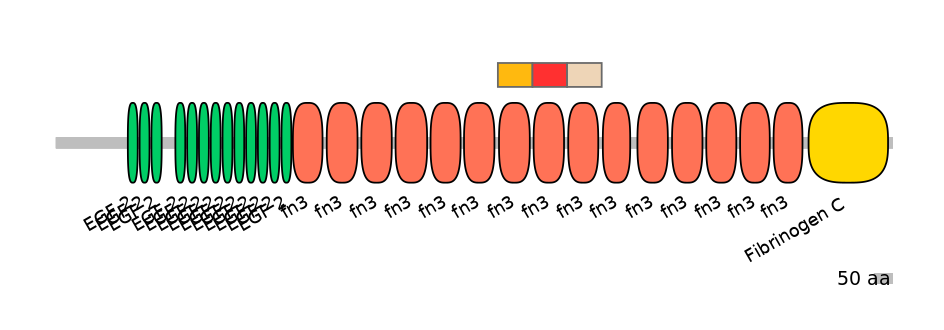
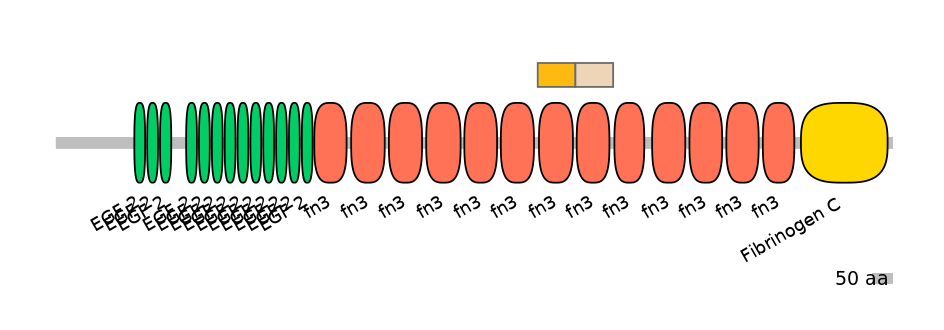
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Mouse
(mm9)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGCACAACCCCTCTCTCTGTT
R:
AGGCCTCAGCAGAGAGTACTG
Band lengths:
301-574
Functional annotations
There are 5 annotated functions for this event
PMID: 7499434
The small splice variant of tenascin-C (TNC) has eight fibronectin type III (FN3) domains (it lacks all alternatively splice exons, including: HsaEX0066148, HsaEX0066149, HsaEX0066151, and HsaEX0066152). The major large splice variant has three (in chicken) or seven (in human) additional FN3 domains (encoded by the alternative exons A1 (HsaEX0066148), A2, A3 (HsaEX0066149), A4, B,C (HsaEX0066151), D (HsaEX0066152)) inserted between domains five and six. Chiquet-Ehrismann et al. (Chiquet-Ehrismann, R., Matsuoka, Y., Hofer, U., Spring, J., Bernasconi, C., and Chiquet, M. (1991, Cell Regul. 2, 927-938) demonstrated that the small variant bound preferentially to fibronectin in enzyme-linked immunosorbent assay, and only the small variant was incorporated into the matrix by cultures of chicken fibroblasts. In this paper the authors have studied human tenascin-C, and confirmed that the small variant binds preferentially to purified fibronectin and to fibronectin-containing extracellular matrix. Thus this differential binding appears to be conserved across vertebrate species. Using bacterial expression proteins, they mapped the major binding site to the third FN3 domain of TNC. Consistent with this mapping, a monoclonal antibody against an epitope in this domain did not stain TNC segments bound to cell culture matrix fibrils. The enhanced binding of the small TN variant suggests the existence of another, weak binding site probably in FN3 domains 6-8, which is only positioned to bind fibronectin in the small splice variant. This binding of domains 6-8 may involve a third molecule present in matrix fibrils, as the enhanced binding of small TN was much more prominent to matrix fibrils than to purified fibronectin. Note, the function of individual exons were not examined.
PMID: 10367733
This study investigated the impact of cellular environment on the neurite outgrowth promoting properties of the alternatively spliced fibronectin type-III region (fnA-D) of tenascin-C. The effects of the A1-A4 region (consists of exon A1 (HsaEX0066148), A2, A3 (HsaEX0066149), and A4), B-D region (consists of exon B, exon C (HsaEX0066151), and exon D (HsaEX0066152) and the entire region A-D (all the mentioned exons) were examined. FnA-D promoted neurite outgrowth in vitro when bound to the surface of BHK cells or cerebral cortical astrocytes, but the absolute increase was greater on astrocytes. In addition, different neurite outgrowth promoting sites were revealed within fnA-D bound to the two cellular substrates. FnA-D also promoted neurite outgrowth as a soluble ligand; however, the actions of soluble fnA-D were not affected by cell type. It was hypothesized that different mechanisms of cellular binding can alter the growth promoting actions of bound fnA-D. The authors found that fnA-D utilizes two distinct sequences to bind to the BHK cell surface as opposed to the BHK extracellular matrix. In contrast, only one of these sequences is utilized to bind to the astrocyte matrix as opposed to the astrocyte surface. Furthermore, Scatchard analysis indicated two types of receptors for fnA-D on BHK cells and only one type on astrocytes. These results suggest that active sites for neurite outgrowth within fnA-D are differentially revealed depending on cell-specific fnA-D binding sites.
PMID: 10493745
Tenascin-C has been implicated in regulation of both neurite outgrowth and neurite guidance. A particular region of tenascin-C has powerful neurite outgrowth-promoting actions in vitro. This region consists of the alternatively spliced fibronectin type-III (FN-III) repeats A?D and is abbreviated fnA-D. The purpose of this study was to investigate whether fnA-D also provides neurite guidance cues and whether the same or different sequences mediate outgrowth and guidance. The examined exons were A1 (HsaEX0066148), A2, A3 (HsaEX0066149), A4, C (HsaEX0066151), and D (HsaEX0066152). The authors developed an assay to quantify neurite behavior at sharp substrate boundaries and found that neurites demonstrated a strong preference for fnA-D when given a choice at a poly-l-lysine?fnA-D interface, even when fnA-D was intermingled with otherwise repellant molecules. Furthermore, neurites preferred cells that overexpressed the largest but not the smallest tenascin-C splice variant when given a choice between control cells and cells transfected with tenascin-C. The permissive guidance cues of large tenascin-C expressed by cells were mapped to fnA-D. Using a combination of recombinant proteins corresponding to specific alternatively spliced FN-III domains and monoclonal antibodies against neurite outgrowth-promoting sites, the authors demonstrated that neurite outgrowth and guidance were facilitated by distinct sequences within fnA-D.
PMID: 11714809
[Likely mapping: domain TnFnIII A3; PMID:25482829]. Together, these 6-8 exons the encode minimal region of tenascin-C that can inhibit T cell activation. Recombinant fragments corresponding to defined regions of the molecule were tested for their ability to inhibit in vitro activation of human peripheral blood T cells induced by anti-CD3 mAbs in combination with fibronectin or IL-2. A recombinant protein encompassing the alternatively spliced fibronectin type III domains of tenascin-C (TnFnIII A1-3 and B-D) vigorously inhibited both early and late lymphocyte activation events including activation-induced TCR/CD8 down-modulation, cytokine production, and DNA synthesis. In agreement with this, full length recombinant tenascin-C containing the alternatively spliced region suppressed T cell activation, whereas tenascin-C lacking this region did not. Using a series of smaller fragments and deletion mutants issued from this region, the authors have identified the TnFnIII A1A2 domain as the minimal region suppressing T cell activation. Single TnFnIII A1 or A2 domains were no longer inhibitory, while maximal inhibition required the presence of the TnFnIII A3 domain.
PMID: 19405959
The stromal microenvironment has a profound influence on tumour cell behaviour. In tumours, the extracellular matrix (ECM) composition differs from normal tissue and allows novel interactions to influence tumour cell function. The ECM protein tenascin-C (TNC) is frequently up-regulated in breast cancer and the same authors have previously identified two novel isoforms - one containing exon 16 (TNC-16) (the exon is also called exon D (HsaEX0066152) and one containing exons 14 (exon B) plus 16 (TNC-14/16). The present study has analysed the functional significance of this altered TNC isoform profile in breast cancer. TNC-16 and TNC-14/16 splice variants were generated using PCR-ligation and over-expressed in breast cancer cells (MCF-7, T47D, MDA-MD-231, MDA-MB-468, GI101) and human fibroblasts. The effects of these variants on tumour cell invasion and proliferation were measured and compared with the effects of the large (TNC-L) (containing alternatively spliced exons A1 (HsaEX0066148), A2, A3 (HsaEX0066149), A4, B, C (HsaEX0066151), D) and fully spliced small (TNC-S) isoforms (skips all cassette exons). TNC-16 and TNC-14/16 significantly enhanced tumour cell proliferation (P < 0.05) and invasion, both directly (P < 0.01) and as a response to transfected fibroblast expression (P < 0.05) with this effect being dependent on tumour cell interaction with TNC, because TNC-blocking antibodies abrogated these responses. An analysis of 19 matrix metalloproteinases (MMPs) and tissue inhibitor of matrix metalloproteinases 1 to 4 (TIMP 1 to 4) revealed that TNC up-regulated expression of MMP-13 and TIMP-3 two to four fold relative to vector, and invasion was reduced in the presence of MMP inhibitor GM6001. However, this effect was not isoform-specific but was elicited equally by all TNC isoforms.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development