HsaEX6062710 @ hg38
Exon Skipping
Gene
ENSG00000041982 | TNC
Description
tenascin C [Source:HGNC Symbol;Acc:HGNC:5318]
Coordinates
chr9:115062917-115064919:-
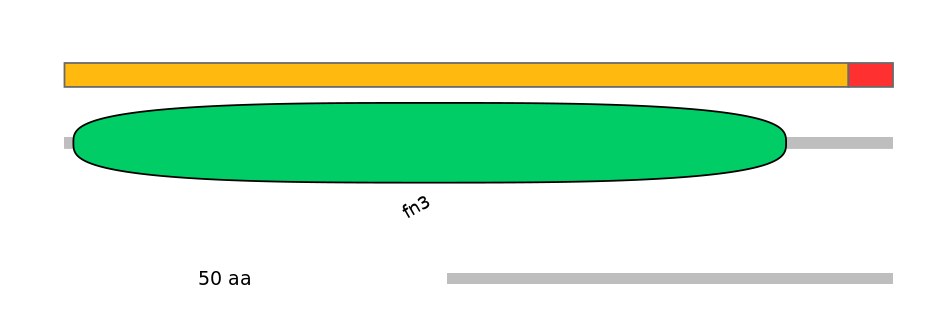
Coord C1 exon
chr9:115064647-115064919
Coord A exon
chr9:115063796-115063961
Coord C2 exon
chr9:115062917-115063189
Length
166 bp
Sequences
Splice sites
3' ss Seq
ATGAGTACTTTTTCATTCAGGTG
3' ss Score
7.13
5' ss Seq
CAGGTATTC
5' ss Score
6.49
Exon sequences
Seq C1 exon
AACAAGCCCCTGAGCTGGAAAACCTCACCGTGACTGAGGTTGGCTGGGATGGCCTCAGACTCAACTGGACCGCAGCTGACCAGGCCTATGAGCACTTTATCATTCAGGTGCAGGAGGCCAACAAGGTGGAGGCAGCTCGGAACCTCACCGTGCCTGGCAGCCTTCGGGCTGTGGACATACCGGGCCTCAAGGCTGCTACGCCTTATACAGTCTCCATCTATGGGGTGATCCAGGGCTATAGAACACCAGTGCTCTCTGCTGAGGCCTCCACAG
Seq A exon
GTGCAGGAGGCTGACACAGTAGAGGCAGCCCAGAACCTCACCGTCCCAGGAGGACTGAGGTCCACAGACCTGCCTGGGCTCAAAGCAGCCACTCATTATACCATCACCATCCGCGGGGTCACTCAGGACTTCAGCACAACCCCTCTCTCTGTTGAAGTCTTGACAG
Seq C2 exon
AGGAGGTTCCAGATATGGGAAACCTCACAGTGACCGAGGTTAGCTGGGATGCTCTCAGACTGAACTGGACCACGCCAGATGGAACCTATGACCAGTTTACTATTCAGGTCCAGGAGGCTGACCAGGTGGAAGAGGCTCACAATCTCACGGTTCCTGGCAGCCTGCGTTCCATGGAAATCCCAGGCCTCAGGGCTGGCACTCCTTACACAGTCACCCTGCACGGCGAGGTCAGGGGCCACAGCACTCGACCCCTTGCTGTAGAGGTCGTCACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000041982-'19-27,'19-26,20-27
Average complexity
C3*
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.099 A=0.099 C2=0.207
Domain overlap (PFAM):
C1:
PF0004116=fn3=WD(100=88.0)
A:
NO
C2:
PF0004116=fn3=WD(100=88.0)
Main Inclusion Isoform:
ENST00000544972fB31571
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAACACCAGTGCTCTCTGCTG
R:
GACTGTGTAAGGAGTGCCAGC
Band lengths:
245-411
Functional annotations
There are 1 annotated functions for this event
PMID: 11714809
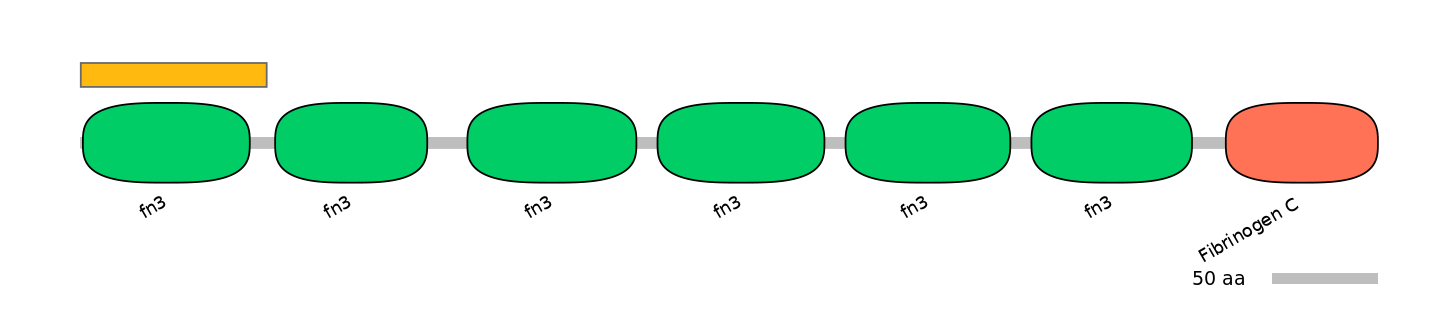
[Likely mapping: domain TnFnIII A2; PMID:25482829]. Together, these 6-8 alternative exon array the encode minimal region of tenascin-C that can inhibit T cell activation. Recombinant fragments corresponding to defined regions of the molecule were tested for their ability to inhibit in vitro activation of human peripheral blood T cells induced by anti-CD3 mAbs in combination with fibronectin or IL-2. A recombinant protein encompassing the alternatively spliced fibronectin type III domains of tenascin-C (TnFnIII A1-3 and B-D) vigorously inhibited both early and late lymphocyte activation events including activation-induced TCR/CD8 down-modulation, cytokine production, and DNA synthesis. In agreement with this, full length recombinant tenascin-C containing the alternatively spliced region suppressed T cell activation, whereas tenascin-C lacking this region did not. Using a series of smaller fragments and deletion mutants issued from this region, the authors have identified the TnFnIII A1A2 domain as the minimal region suppressing T cell activation. Single TnFnIII A1 or A2 domains were no longer inhibitory, while maximal inhibition required the presence of the TnFnIII A3 domain.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains