MmuEX0048238 @ mm9
Exon Skipping
Gene
ENSMUSG00000028364 | Tnc
Description
tenascin C [Source:MGI Symbol;Acc:MGI:101922]
Coordinates
chr4:63637480-63667543:-
Coord C1 exon
chr4:63667280-63667543
Coord A exon
chr4:63660947-63661219
Coord C2 exon
chr4:63637480-63637602
Length
273 bp
Sequences
Splice sites
3' ss Seq
CATTCTTTGTCCTTCTCTAGGGA
3' ss Score
11.07
5' ss Seq
CAGGTATTT
5' ss Score
7.51
Exon sequences
Seq C1 exon
AAATTGATGCACCCAAGGACTTACGGGTGTCTGAAACCACACAAGACAGTCTGACGTTTTTCTGGACGACACCCCTGGCCAAGTTTGATCGTTACCGCCTCAACTACAGCCTCCCCACAGGCCAGTCGATGGAAGTCCAGCTGCCAAAGGATGCCACCTCCCATGTCCTGACAGACCTGGAGCCAGGGCAAGAATACACTGTTCTCCTCATTGCTGAGAAGGGCAGACACAAGAGCAAGCCTGCACGTGTGAAGGCATCCACGG
Seq A exon
GGACAACTCCCAATCTGGGAGAGGTCACTGTGGCCGAGGTGGGCTGGGATGCCCTCACGCTCAACTGGACTGCTCCAGAAGGAGCCTATAAGAACTTTTTCATTCAGGTGCTAGAGGCTGACACGACCCAGACTGTCCAGAACCTCACAGTCCCAGGAGGACTGAGGTCAGTGGACCTGCCTGGGCTCAAAGCAGCCACCCGCTACTACATCACCCTTCGAGGGGTCACCCAGGACTTCGGCACGGCCCCTCTCTCTGTTGAGGTCTTGACAG
Seq C2 exon
CCATGGGTTCTCCGAAGGAAATCATGTTCTCAGACATCACTGAAAATGCAGCCACAGTCAGCTGGAGGGCACCTACTGCTCAGGTGGAGAGTTTCCGGATCACTTATGTACCTATGACAGGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000028364_MULTIEX1-2/6=C1-C2
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.404 A=0.076 C2=0.031
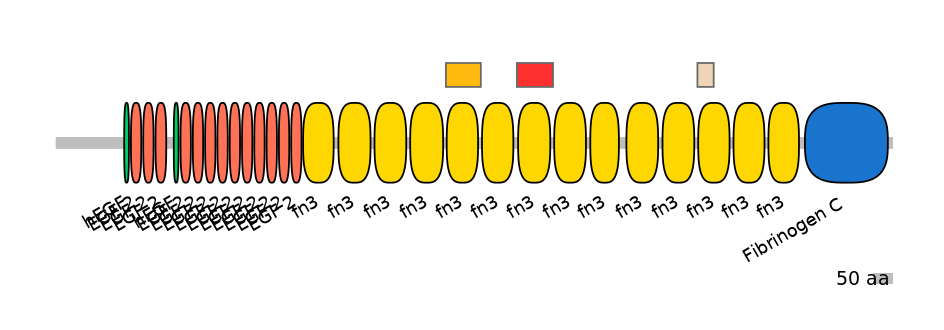
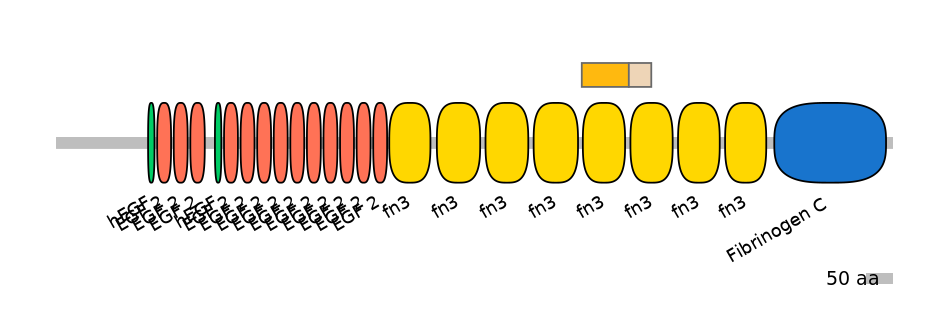
Domain overlap (PFAM):
C1:
PF0004116=fn3=WD(100=89.9)
A:
PF0004116=fn3=WD(100=89.1)
C2:
PF0004116=fn3=PU(48.8=92.9)
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGGGTGTCTGAAACCACACAA
R:
AGCTGACTGTGGCTGCATTTT
Band lengths:
304-577
Functional annotations
There are 1 annotated functions for this event
PMID: 11714809
[Likely mapping: domain TnFnIII A2; PMID:25482829]. Together, these 6-8 alternative exon array the encode minimal region of tenascin-C that can inhibit T cell activation. Recombinant fragments corresponding to defined regions of the molecule were tested for their ability to inhibit in vitro activation of human peripheral blood T cells induced by anti-CD3 mAbs in combination with fibronectin or IL-2. A recombinant protein encompassing the alternatively spliced fibronectin type III domains of tenascin-C (TnFnIII A1-3 and B-D) vigorously inhibited both early and late lymphocyte activation events including activation-induced TCR/CD8 down-modulation, cytokine production, and DNA synthesis. In agreement with this, full length recombinant tenascin-C containing the alternatively spliced region suppressed T cell activation, whereas tenascin-C lacking this region did not. Using a series of smaller fragments and deletion mutants issued from this region, the authors have identified the TnFnIII A1A2 domain as the minimal region suppressing T cell activation. Single TnFnIII A1 or A2 domains were no longer inhibitory, while maximal inhibition required the presence of the TnFnIII A3 domain.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: