MmuEX0017494 @ mm9
Exon Skipping
Gene
ENSMUSG00000011831 | Evi5
Description
ecotropic viral integration site 5 [Source:MGI Symbol;Acc:MGI:104736]
Coordinates
chr5:108271090-108304126:-
Coord C1 exon
chr5:108304008-108304126
Coord A exon
chr5:108301620-108301674
Coord C2 exon
chr5:108271090-108271319
Length
55 bp
Sequences
Splice sites
3' ss Seq
GCGATGCTCCTGTCTTACAGTCA
3' ss Score
7.45
5' ss Seq
CCGGTAAGT
5' ss Score
10.91
Exon sequences
Seq C1 exon
AAACACTCCCTATGAGTCAGCTGTAAATTCAATTCAAATCCCCAGTCTCCTAGTCTCCTCCTCTCATCATGGTTACCACCAAAATGACTGCTGCCTTTAGAAACCCTAATAGGAGACAG
Seq A exon
TCAGTGGATACGGTGAAGGAAAAGTCCTGAGCGTGAAGCCATTTTTAGAACCCCG
Seq C2 exon
GTGGCGACAGACAAGGTGGCAGAAAAGCTGAGTTCTACTCTCTCATGGGTGAAGAACACAGTATCGCATACAGTCAGTCAGATGGCCAGTCAGGTGGCAAGTCCATCTGCCTCACTACACACTACATCCTCATCTACCACACTGTCAACACCAACACAGTCACCATCTTCCCCATCAAAGTTGAGTCCAGACGACTTAGAACTCCTGGCTAAGCTGGAGGAACAAAACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000011831_CASSETTE3
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
5' UTR
No structure available
Features
Disorder rate (Iupred):
C1=0.961 A=1.000 C2=0.759
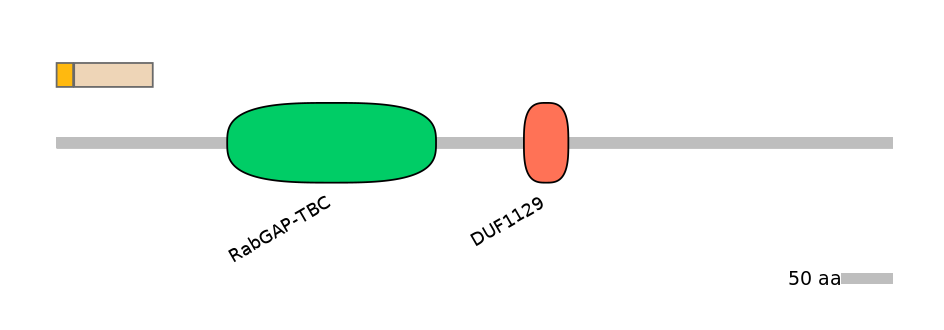
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Human
(hg38)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCACCAAAATGACTGCTGCCT
R:
ACTGGCCATCTGACTGACTGT
Band lengths:
134-189
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: