MmuEX0041127 @ mm10
Exon Skipping
Gene
ENSMUSG00000035236 | Scai
Description
suppressor of cancer cell invasion [Source:MGI Symbol;Acc:MGI:2443716]
Coordinates
chr2:39132955-39190033:-
Coord C1 exon
chr2:39189989-39190033
Coord A exon
chr2:39150314-39150414
Coord C2 exon
chr2:39132955-39133086
Length
101 bp
Sequences
Splice sites
3' ss Seq
ACTTATTTTTTTTCACCCAGATT
3' ss Score
7.83
5' ss Seq
ATGGTAAAT
5' ss Score
6.41
Exon sequences
Seq C1 exon
ACTGTCTGGCACAGTGGAGAAACCACCCCGAAAACGGAAAAGCAG
Seq A exon
ATTTCTTAAGCAACTACTGTCTGACTACTGTCTGGAGATGAAGAAATTTTAAAACGACGTCTCTCTCCTCCTGGAGTTTTTGTTATATCTGGAGATAGATG
Seq C2 exon
GACTGAATTTACTCTTAAAGAAACTATGTCCTCTGGAGGTGCTGAAGATGACATCCCACAGGGAGAGAGGAAGACAGTTACAGATTTTTGTTATCTTTTGGATAAATCCAAGCAACTGTTCAATGGCTTAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000035236_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.973 A=0.000 C2=0.444
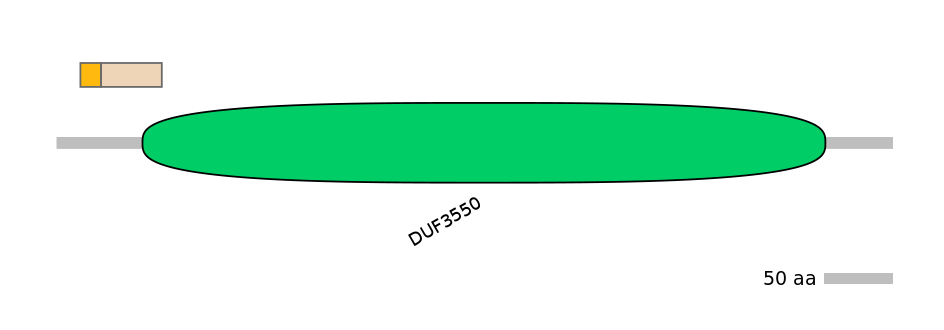
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF120703=DUF3550=PU(2.8=31.1)
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGCACAGTGGAGAAACC
R:
TTAAGCCATTGAACAGTTGCTTGG
Band lengths:
170-271
Functional annotations
There are 1 annotated functions for this event
PMID: 32736682
This event
This study identified novel rat Scai variants 2 and 3 and characterized their functions in Neuro-2a cells. The novel Scai variants 2 and 3 contain unique exons that possess stop codons and therefore encode shorter proteins compared with the full-length Scai variant 1. SCAI variants 2 and 3 possess a nuclear localization signal, but do not have an MRTF-binding site. Immunostaining of green fluorescent protein (GFP)-tagged SCAI variants revealed a nuclear localization of variant 1, whereas localization of variants 2 and 3 was throughout the cytoplasm and nucleus, suggesting that other nuclear localization signals, which act in Neuro-2a cells, exist in SCAI. All three SCAI variants suppressed the neuron-like morphological change of Neuro-2a cells induced by a Rho effector, constitutively active mDia; however, the suppressive effects of variants 2 and 3 were weaker than that of full-length SCAI variant 1, indicating that the SCAI-mediated change toward a neuronal morphology appeared to be consistent with their nuclear localization. These findings indicate that generation of multiple SCAI splice variants fines-tune neuronal morphology.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types