MmuEX6028380 @ mm10
Exon Skipping
Gene
ENSMUSG00000026471 | Mr1
Description
major histocompatibility complex, class I-related [Source:MGI Symbol;Acc:MGI:1195463]
Coordinates
chr1:155130633-155136850:-
Coord C1 exon
chr1:155136575-155136850
Coord A exon
chr1:155132360-155132635
Coord C2 exon
chr1:155130633-155130731
Length
276 bp
Sequences
Splice sites
3' ss Seq
CTGTGGCTGCTTCTTTCTAGAGC
3' ss Score
8.04
5' ss Seq
GGGGTTAGT
5' ss Score
2.75
Exon sequences
Seq C1 exon
GGCTTCACACCTACCAGAGAATGATTGGCTGTGAGTTGCTAGAAGATGGCAGCACCACAGGGTTTCTCCAGTATGCATATGATGGACAAGATTTCATCATCTTCAATAAAGACACCCTCTCCTGGTTGGCCATGGATTATGTGGCTCACATCACCAAGCAAGCATGGGAGGCCAACCTGCATGAGTTGCAATACCAAAAGAACTGGCTGGAAGAAGAATGCATTGCCTGGCTAAAGAGGTTCTTGGAATATGGAAGAGATACCCTAGAAAGAACAG
Seq A exon
AGCATCCAGTGGTAAGAACAACTCGAAAGGAAACTTTTCCAGGGATTACAACTTTCTTCTGCAGAGCTCATGGCTTCTACCCACCAGAAATTTCCATGACATGGATGAAAAATGGGGAAGAAATTGCCCAAGAAGTGGATTACGGAGGGGTACTTCCCAGCGGGGATGGAACCTATCAAACGTGGCTGTCAGTTAATCTGGATCCTCAGAGCAATGATGTTTATTCTTGTCATGTGGAGCACTGTGGTCGCCAAATGGTTCTGGAGGCCCCTCGGG
Seq C2 exon
AATCAGGGGACATCCTTCGAGTGAGCACGATCTCTGGAACCACAATTCTCATCATCGCCCTGGCTGGAGTTGGTGTTCTGATCTGGAGAAGGTCACAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000026471-'7-6,'7-4,9-6
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
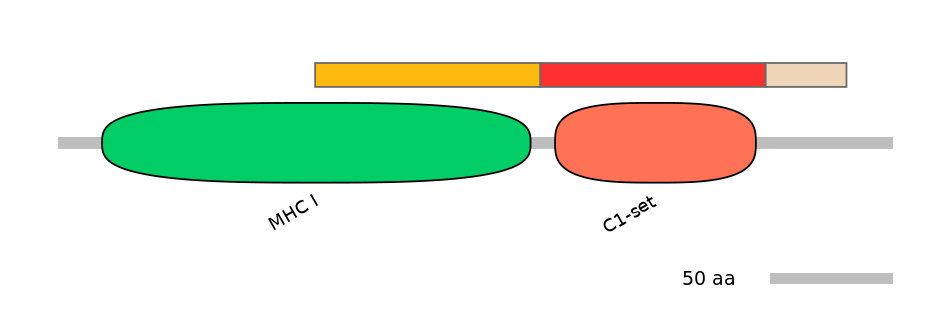
Features
Disorder rate (Iupred):
C1=0.009 A=0.065 C2=0.000
Domain overlap (PFAM):
C1:
PF0012913=MHC_I=PD(50.0=94.6)
A:
PF0765410=C1-set=WD(100=89.2)
C2:
NO
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCACAGGGTTTCTCCAGTATGC
R:
CAGAACACCAACTCCAGCCAG
Band lengths:
302-578
Functional annotations
There are 1 annotated functions for this event
PMID: 32963314
The full-length isoform, MR1A, can activate Mucosal Associated Invariant T (MAIT) cells. Using a transcriptomic analysis in conjunction with qPCR, the authors find that that MR1A and MR1B (skipping of HsaEX0040046) transcripts are widely expressed. However only MR1A can present mycobacterial antigen to MAIT cells. Coexpression of MR1B with MR1A decreases MAIT cell activation following bacterial infection. Additionally, expression of MR1B prior to MR1A lowers total MR1A abundance, suggesting competition between MR1A and MR1B for either ligands or chaperones required for folding and/or trafficking. Finally, the authors evaluated CD4/CD8 double positive thymocytes expressing surface MR1. Relative expression of MR1A/MR1B transcript is associated with the prevalence of MR1 + CD4/CD8 cells in the thymus.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types