RnoEX0055530 @ rn6
Exon Skipping
Gene
ENSRNOG00000003522 | Mr1
Description
major histocompatibility complex, class I-related [Source:RGD Symbol;Acc:1593291]
Coordinates
chr13:72773739-72779981:-
Coord C1 exon
chr13:72779706-72779981
Coord A exon
chr13:72775422-72775697
Coord C2 exon
chr13:72773739-72773843
Length
276 bp
Sequences
Splice sites
3' ss Seq
CTGTGGTTGCTTCTCTCTAGAAC
3' ss Score
9.04
5' ss Seq
AGGGTAAGT
5' ss Score
10.45
Exon sequences
Seq C1 exon
GGCTTCACACCTACCAGAGAATGATCGGTTGTGAGTTGCTGGAAGACGGCAGCACCACAGGGTTTCTCCAGTATGCATATGATGGACAAGATTTCATTGTCTTCGATAAAGACACCCTCTCCTGGCTGGCTATGGATAATGTGGCTCACATCACCAAGCGAGCATGGGAGGCCAACCTGCATGAGTTGCAATACCAAAAGAACTGGCTAGAAGAAGAGTGCATTGCCTGGCTAAAGAGGTTCTTGGAGTATGGAAGTGATGCCCTAGAAAGAACAG
Seq A exon
AACATCCAGTAGTAAGAACAACTCGAAAGGAAACGTTTCCAGGGATTACCACTCTCTTCTGCAGAGCTCATGGCTTCTACCCACCAGAAATTTCCATGATATGGAAGAAAAATGGGGAAGAAATTGTCCAGGAGGTGGATTATGGAGGGGTGCTTCCCAGTGGGGATGGAACCTATCAAATGTGGGTGTCGGTTGATCTGGATCCTCAGACCAAAGACATTTATTCTTGTCATGTGGAGCACTGTGGTCTCCAAATGGTTCTGGAGGCCCCTCAGG
Seq C2 exon
AATCAGGAAACACCCTTCTAGTGGCAAACACGATCTCTGGGACCATAATTCTCATCATTGTCCTGGCTGGAGTTGGTGCCCTGATCTGGAGAAGAAGGTCACGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000003522_MULTIEX1-3/4=1-C2
Average complexity
C1*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.007 A=0.048 C2=0.119
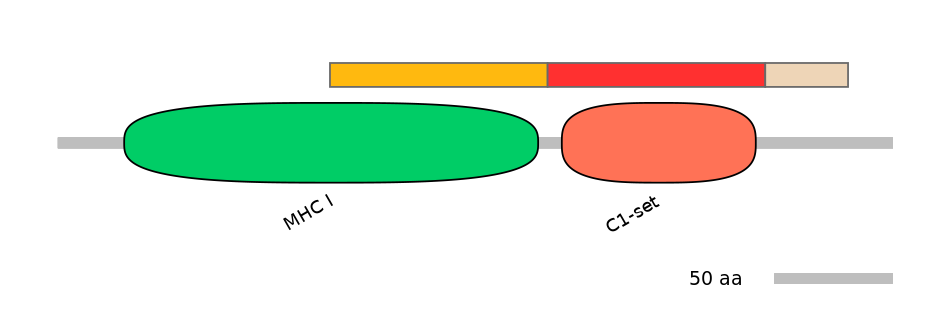
Domain overlap (PFAM):
C1:
PF0012913=MHC_I=PD(50.0=94.6)
A:
PF0765410=C1-set=WD(100=89.2)
C2:
NO
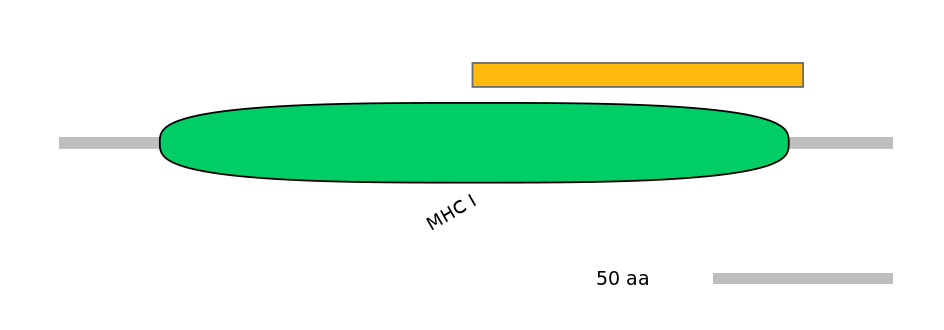
Main Skipping Isoform:
ENSRNOT00000004694fB8929
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGATCGGTTGTGAGTTGCTGG
R:
GTCCCAGAGATCGTGTTTGCC
Band lengths:
298-574
Functional annotations
There are 1 annotated functions for this event
PMID: 32963314
The full-length isoform, MR1A, can activate Mucosal Associated Invariant T (MAIT) cells. Using a transcriptomic analysis in conjunction with qPCR, the authors find that that MR1A and MR1B (skipping of HsaEX0040046) transcripts are widely expressed. However only MR1A can present mycobacterial antigen to MAIT cells. Coexpression of MR1B with MR1A decreases MAIT cell activation following bacterial infection. Additionally, expression of MR1B prior to MR1A lowers total MR1A abundance, suggesting competition between MR1A and MR1B for either ligands or chaperones required for folding and/or trafficking. Finally, the authors evaluated CD4/CD8 double positive thymocytes expressing surface MR1. Relative expression of MR1A/MR1B transcript is associated with the prevalence of MR1 + CD4/CD8 cells in the thymus.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]