MmuINT0145930 @ mm9
Intron Retention
Gene
ENSMUSG00000059316 | Slc27a4
Description
solute carrier family 27 (fatty acid transporter), member 4 [Source:MGI Symbol;Acc:MGI:1347347]
Coordinates
chr2:29668072-29668454:+
Coord C1 exon
chr2:29668072-29668236
Coord A exon
chr2:29668237-29668307
Coord C2 exon
chr2:29668308-29668454
Length
71 bp
Sequences
Splice sites
5' ss Seq
CAGGTATGT
5' ss Score
9.8
3' ss Seq
GTGTACCCTCCTGCTTCCAGGAA
3' ss Score
10.11
Exon sequences
Seq C1 exon
GTGACGTCCTGGTGATGGATGAGCTGGGTTACCTGTACTTCCGAGATCGCACTGGGGACACGTTCCGCTGGAAAGGGGAGAATGTATCTACCACTGAGGTGGAGGGCACACTCAGCCGCCTGCTTCATATGGCAGATGTGGCAGTTTATGGTGTTGAGGTGCCAG
Seq A exon
GTATGTGAGGGGAGGATGGTGAGCATGTCCGAACATTGCCTGCCACTGACTGTGTACCCTCCTGCTTCCAG
Seq C2 exon
GAACTGAAGGCCGAGCAGGAATGGCTGCCGTTGCAAGTCCCATCAGCAACTGTGACCTGGAGAGCTTTGCACAGACCTTGAAAAAGGAGCTGCCTCTGTATGCCCGCCCCATCTTCCTGCGCTTCTTGCCTGAGCTGCACAAGACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000059316-Slc27a4:NM_011989:11
Average complexity
IR-S
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.054 A=NA C2=0.000
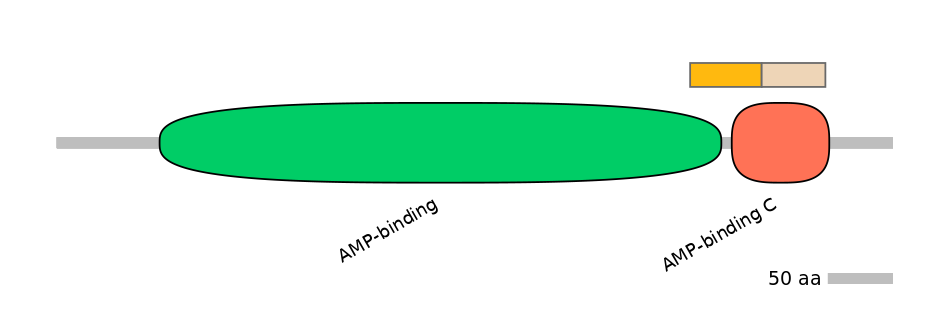
Domain overlap (PFAM):
C1:
PF0050123=AMP-binding=PD(5.5=42.9),PF131931=AMP-binding_C=PU(30.3=41.1)
A:
NA
C2:
PF131931=AMP-binding_C=FE(64.5=100)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTACTTCCGAGATCGCACTGG
R:
CAGTTGCTGATGGGACTTGCA
Band lengths:
183-254
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: