RnoALTD0018803-2/2 @ rn6
Alternative 5'ss
Gene
ENSRNOG00000019680 | Mst1
Description
macrophage stimulating 1 [Source:RGD Symbol;Acc:3114]
Coordinates
chr8:116859445-116859908:+
Coord C1 exon
chr8:116859445-116859563
Coord A exon
chr8:116859564-116859632
Coord C2 exon
chr8:116859713-116859908
Length
69 bp
Sequences
Splice sites
5' ss Seq
GAGGTACTA
5' ss Score
6.36
3' ss Seq
CAATTCCAATTTTTGGGCAGGGC
3' ss Score
4.25
Exon sequences
Seq C1 exon
GTTCCCAGACAAAGCTCTGAAAGACAACTATTGCCGTAATCCGGATGCATCTGAGCGGCCCTGGTGCTACACCACGGACCCGAATGTTGAGCGAGAGTTCTGTGACCTGCCCAGTTGCG
Seq A exon
GTATGCTGCAGGGACAGGGCCTCGGAGGGAACTTGGAAAAAACTGGCAGGCGTGGTTCAACTAGGAGAG
Seq C2 exon
GGCCCAACCTGCCACCGACCACCAAAGGATCCAAGTCACAACAGCGCAACAAGGTCAAGGCTTCGAACTGCTTCCGCGGAAAAGGTGAAGACTATCGAGGCACAACCAATACCACCTCTGCGGGTGTGCCCTGCCAGCGCTGGGATGCGCAGAATCCGCACCAGCACCGCTTTGTGCCGGAGAAATATGCTTGCAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000019680-13-8,14-8-2/2
Average complexity
Alt5
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
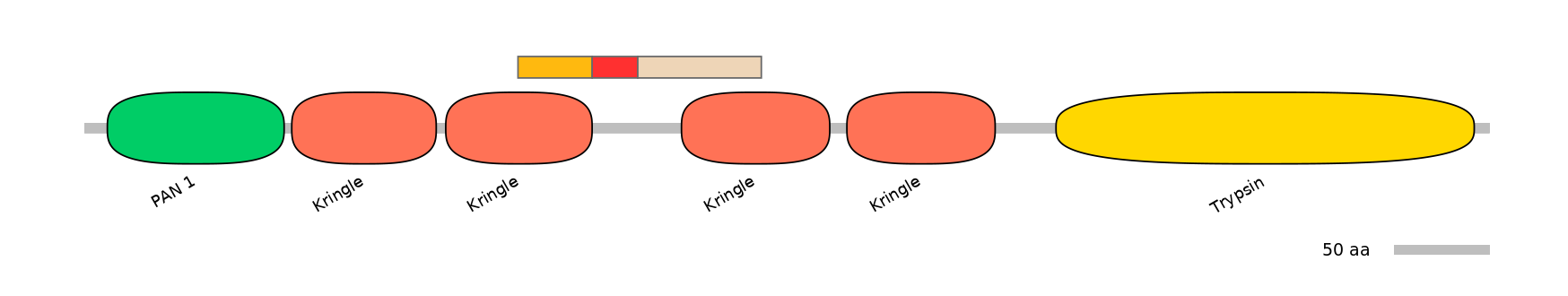
Features
Disorder rate (Iupred):
C1=0.344 A=NA C2=0.470
Domain overlap (PFAM):
C1:
PF0005113=Kringle=PD(50.0=60.9)
A:
NA
C2:
PF0005113=Kringle=PU(53.2=63.6)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TAATCCGGATGCATCTGAGCG
R:
TCTTCACCTTTTCCGCGGAAG
Band lengths:
174-243
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]