RnoEX0009396 @ rn6
Exon Skipping
Gene
ENSRNOG00000053288 | Ank3
Description
ankyrin 3 [Source:RGD Symbol;Acc:620157]
Coordinates
chr20:20385488-20408649:+
Coord C1 exon
chr20:20385488-20385590
Coord A exon
chr20:20403689-20403795
Coord C2 exon
chr20:20408425-20408649
Length
107 bp
Sequences
Splice sites
3' ss Seq
TGAGCCGCTTTGTTTTCCAGCAC
3' ss Score
7.86
5' ss Seq
TGGGTAAGT
5' ss Score
10.24
Exon sequences
Seq C1 exon
CCTTCGCTCCTTCAGTTCGGATAGGTCCTACACCCTGAACAGAAGCTCCTATGCGCGGGACAGCATGATGATCGAGGAGCTTCTGGTGCCATCCAAAGAGCAG
Seq A exon
CACCTTCCATTCACGAGGGAGTTTGACTCCGACTCCCTCAGACACTACAGTTGGGCAGCAGACACACTGGACAATGTGAACCTGGTCTCCAGCCCTGTGCATTCTGG
Seq C2 exon
GTTTCTGGTTAGCTTTATGGTGGACGCGAGAGGGGGCTCCATGCGAGGAAGCCGCCACCACGGGATGCGGATCATCATCCCTCCGCGCAAGTGTACGGCCCCCACCCGCATCACTTGCCGCCTGGTAAAGAGACATAAACTGGCCAACCCACCCCCCATGGTGGAAGGAGAGGGATTAGCCAGTAGGCTGGTAGAAATGGGTCCTGCGGGGGCACAATTTTTAGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000053288_MULTIEX4-2/3=1-C2
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.042 C2=0.218
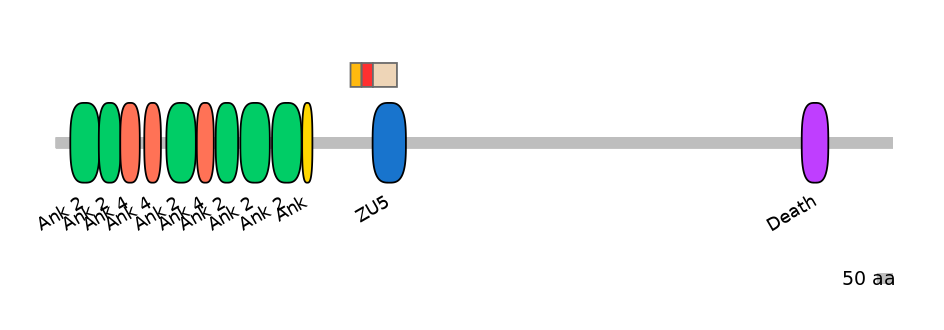
Domain overlap (PFAM):
C1:
NO
A:
PF0079115=ZU5=PU(1.0=2.8)
C2:
PF0079115=ZU5=FE(71.4=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACACCCTGAACAGAAGCTCCT
R:
ATCCCTCTCCTTCCACCATGG
Band lengths:
250-357
Functional annotations
There are 1 annotated functions for this event
PMID: 26024478
The paper describes and annotates all exons to an extreme detail. Wild-type isoform, but not the variants (either skipping 31 or 28-31), bind beta-spectrin. Exon 28 (MmuEX0004926), exon 29 (MmuEX6048924), exon 31 (MmuEX6048923)
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]