RnoEX0018716 @ rn6
Exon Skipping
Gene
ENSRNOG00000003927 | Cd55
Description
CD55 molecule [Source:RGD Symbol;Acc:620651]
Coordinates
chr13:47139046-47147577:-
Coord C1 exon
chr13:47147492-47147577
Coord A exon
chr13:47141852-47142040
Coord C2 exon
chr13:47139046-47139180
Length
189 bp
Sequences
Splice sites
3' ss Seq
ATACTTTGATTCTTTTTAAGCAA
3' ss Score
5.78
5' ss Seq
TAGGTAATC
5' ss Score
5.91
Exon sequences
Seq C1 exon
GTACAAGCTGGTCGGTGTAACTTCTACATTCTGTTCTATTACGGGGAATGCTGTGGACTGGGATGACGTATTTCCAGTGTGCACAA
Seq A exon
CAATTTTTTGTCCAGACCCACCAAAAATCAATAATGGAAGAATTCGAGAGGAAAGCGACTCCTACGCATATAGACAGTCGGTGACCTATTCCTGTGACCAAGGCTTCATTCTGGTTGGAAATTCTTCCGTTCATTGTACTCTGAACGGTGAAGTAGGAGAATGGAGTAGTGCACCACCCAAGTGCATAG
Seq C2 exon
AGAGACACAAGGTCCCAACTAAGACCCCACCAGAAGTTGATATTCCAAGTACAATGCCTCATGAATCCACCTTAATTAATTTTCCAAGTACAAGAGCCCCACTGTCTCATAAACCCACTACAGTTAAAGTTCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000003927_CASSETTE1
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
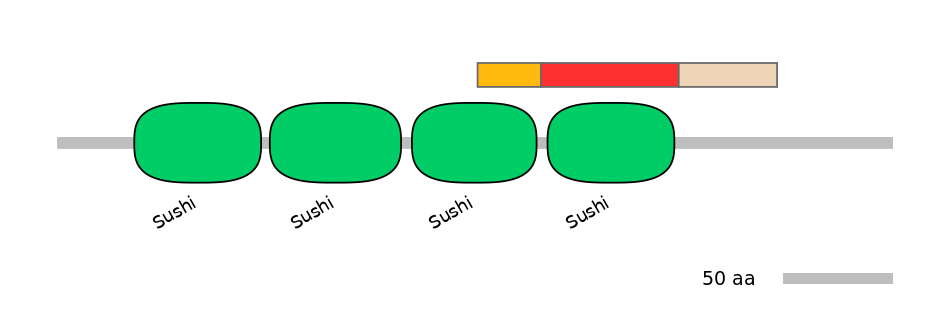
Features
Disorder rate (Iupred):
C1=0.000 A=0.078 C2=0.913
Domain overlap (PFAM):
C1:
PF0008415=Sushi=PD(46.6=90.0)
A:
PF0008415=Sushi=WD(100=92.2)
C2:
NO
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACAAGCTGGTCGGTGTAACTT
R:
CTGGAACTTTAACTGTAGTGGGT
Band lengths:
219-408
Functional annotations
There are 1 annotated functions for this event
PMID: 17007930
Alternative splicing generates either a protein with a GPI anchor or with a transmembrane domain. Different localizations shown by immunohistochemistry.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]