RnoEX0020324 @ rn6
Exon Skipping
Gene
ENSRNOG00000052990 | Cers5
Description
ceramide synthase 5 [Source:RGD Symbol;Acc:1311597]
Coordinates
chr7:141386934-141389631:-
Coord C1 exon
chr7:141389475-141389631
Coord A exon
chr7:141388534-141388641
Coord C2 exon
chr7:141386934-141387772
Length
108 bp
Sequences
Splice sites
3' ss Seq
TGCGTTTCTCTCCTGACTAGGTG
3' ss Score
10.35
5' ss Seq
CATGTAAGT
5' ss Score
8.31
Exon sequences
Seq C1 exon
GATTCTGAACACAACCCTCTTTGAGAGCTGGGAGATTATTGGGCCCTTTCCTTCCTGGTGGCTCTTCAATGGCCTCCTCCTGATCCTACAGATGTTGCATGTCATCTGGTCCTACCTAATCGCACGAACTGCTTTTAAAGCCTTGGTCCGGGGCAAG
Seq A exon
GTGACCTACCCAGGAGGGATTAGCCCCGGCTCTGTACCCTCTGCTCTTTTCTCACCTCCTTCCTCTTTTCTATGCCTGGTGGCAGCTGGACAGTCCCATCCTCTGCAT
Seq C2 exon
GTGTCTAAGGATGACCGCAGTGATGTAGAAAGCAGCTCAGAGGAAGATGAGACCACACACACGAACAACCTCTCTGGCAGCAGTTCCAGCAACGGTGCCAACTGCATGAACGGCCGCATGGGAGGCAGCCACTTGGCTGAGGAGCAGGGGACTTGTAAAGCTGCTGGCAACCTACACTTTGGGGCCTCCCCACGTATACACTCCTGTGACTAGAGGGACCGCCAGGCACTGAGGAGAATCTGAGAAACAAATGTTTTTGTGTTTTAAAGAAACTCTGCCATTTTGTATTTAATGCCCTGGTCCTTCAGTGACATCACTGGCTCTTTGTCTGTGTGTCTGTGCGTCTGTACATGTGCATCATGCATGTGGATGAGCGTCATCCACTAGGTTGGGCACAAGGCTGGATTACTAGTCAGTGTCTGGTCACTTCTGAATCACCTCCGGTCCAGCGTTCGCTGTATCTTGAGTTAAGCTGTCTCCCAACCAGCTGTGTCCTCTTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000052990_MULTIEX1-4/4=3-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.443
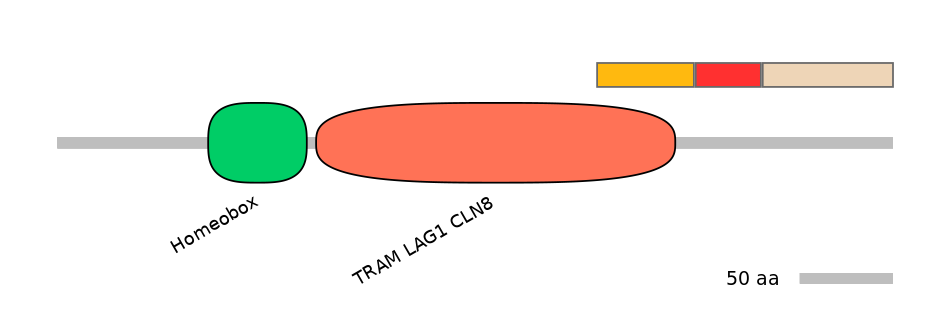
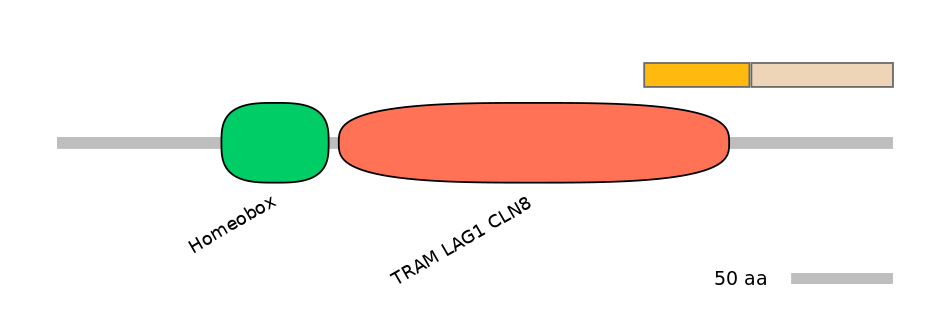
Domain overlap (PFAM):
C1:
PF0379811=TRAM_LAG1_CLN8=PD(21.6=79.2)
A:
NO
C2:
NO
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGCCTCCTCCTGATCCTACAG
R:
GCTTTACAAGTCCCCTGCTCC
Band lengths:
248-356
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]