RnoEX0021739 @ rn6
Exon Skipping
Gene
ENSRNOG00000054251 | Clec7a
Description
C-type lectin domain containing 7A [Source:RGD Symbol;Acc:1565140]
Coordinates
chr4:163222641-163225245:-
Coord C1 exon
chr4:163225147-163225245
Coord A exon
chr4:163224007-163224141
Coord C2 exon
chr4:163222641-163222792
Length
135 bp
Sequences
Splice sites
3' ss Seq
GCTGTTCGTGACATATGAAGCAT
3' ss Score
1.15
5' ss Seq
CAGGCAAGG
5' ss Score
3.33
Exon sequences
Seq C1 exon
GAAGCCCAGCTCCATCTTCTCGTTGGCGGTCCATTGCGGTAGCTTTAGGAATCCTGTGCTTGCTCACAGTAGTGGTCGCAGCAGTGCTGGGTGCCCTAG
Seq A exon
CATTTCGGAGATTCAATTCAGGGAGATATCCAGAGGAGAAAGACAACTTCCCATCAAGAAATAAGGAGAACCACAAACCCACAGAACCATCTTTAGATGAGAAGGTGGCTCCCTCCAAGGCATCCCAAACTACAG
Seq C2 exon
GCGTCTTTTCTGGACCTTGCCTTCCCAATTGGATCATGCATGCCAAGAGCTGTTATCTATTTAGCTTCTCAGAAAATTCCTGGTATGGAAGTAGGAGACACTGTTCCCAGCTAGGAGCTCATCTATTGAAGATAGACAATGCAAAAGAATTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000054251_CASSETTE2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.088 A=0.609 C2=0.000
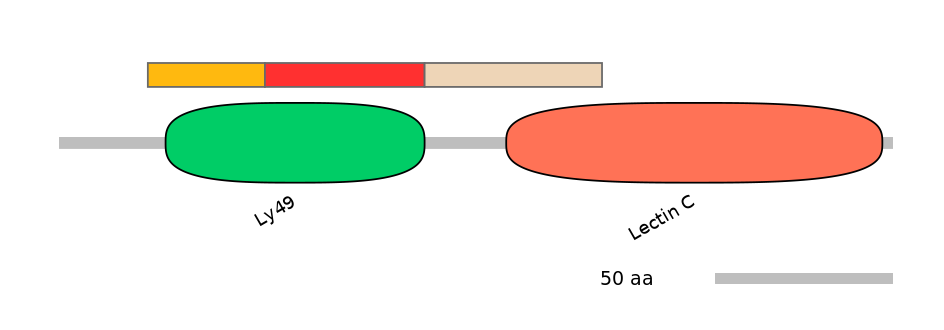
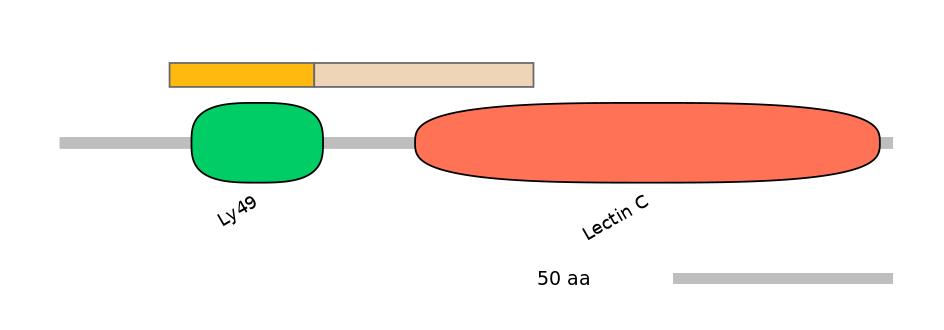
Domain overlap (PFAM):
C1:
PF083915=Ly49=PU(90.3=82.4)
A:
PF083915=Ly49=FE(60.8=100)
C2:
PF083915=Ly49=PD(6.5=3.9),PF0005916=Lectin_C=PU(25.2=52.9)
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAAGCCCAGCTCCATCTTCTC
R:
TGCATTGTCTATCTTCAATAGATGAG
Band lengths:
242-377
Functional annotations
There are 2 annotated functions for this event
PMID: 11567029
[Negative results]. The two major isoforms differ by the presence of a stalk region separating the carbohydrate recognition domain from the transmembrane region and are the only isoforms that are functional for beta-glucan binding.
PMID: 16622020
Overexpression of dectin1-1B results in significantly lower zymosan association compared to the overexpression of dectin-1A at 4?C, 25?C, and 30?C but similar zymosan association at 37?C.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]