RnoEX0023923 @ rn6
Exon Skipping
Gene
ENSRNOG00000050052 | Cox19
Description
COX19 cytochrome c oxidase assembly factor [Source:RGD Symbol;Acc:1305631]
Coordinates
chr12:17395624-17398355:+
Coord C1 exon
chr12:17395624-17395733
Coord A exon
chr12:17396537-17396790
Coord C2 exon
chr12:17398244-17398355
Length
254 bp
Sequences
Splice sites
3' ss Seq
GCCTCCTGGTGCCCGTACAGGAC
3' ss Score
7.27
5' ss Seq
GAGGTACAG
5' ss Score
6.62
Exon sequences
Seq C1 exon
GCCGCTCTCCGCTCTTGATTTCGCCGTGATGTCGACCGCAATGAACTTCGGGACCAAAAGCTTCCAGCCGCGGCCCCCAGACAAAGGCAGCTTCCCGCTAGACCACTTCG
Seq A exon
GACAGGACACGCCTCCCTCCGATGCTCTGGGTCTGCAGTGAACCGTTAGGCAAAGAACTGTTTTCAGAAAGCTTTCCGATGTCTAATACTGGGGCTGAAAGCCAGGGCTTGGCACATGCTGGGTGAGTGCTTCACCTCAGAGCCACACCTGCAGCCCAGAGGATTCCCAGTCACCATTCCGAAGTCTACAGTTGACCTGTCCTTCGGGAAACCGAAACGTGTAAAAGAAAAAAACAACTGGGAGAGAGTTGGAG
Seq C2 exon
GTGAGTGTAAAAGCTTTAAGGAAAAATTCATGAAGTGTCTCCGCGACAAGAACTATGAAAATGCTCTGTGCAGAAATGAATCTAAAGAGTATTTAATGTGCAGGATGCAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000050052_MULTIEX1-1/2=C1-2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.607 A=1.000 C2=0.000
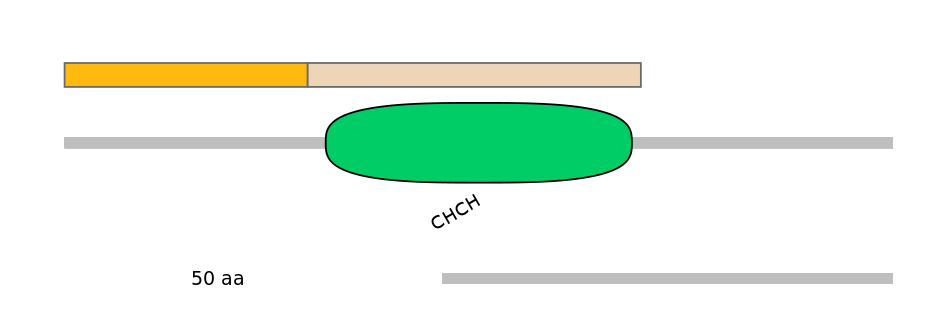
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF067478=CHCH=WD(100=92.1)
Main Inclusion Isoform:
ENSRNOT00000072509fB3484
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTCTCCGCTCTTGATTTCGCC
R:
GCATCCTGCACATTAAATACTCTT
Band lengths:
214-468
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]