BtaEX0017371 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000003829 | HPN
Description
Bos taurus hepsin (HPN), mRNA. [Source:RefSeq mRNA;Acc:NM_001080241]
Coordinates
chr18:45979118-45985453:+
Coord C1 exon
chr18:45979118-45979219
Coord A exon
chr18:45979308-45979349
Coord C2 exon
chr18:45985324-45985453
Length
42 bp
Sequences
Splice sites
3' ss Seq
CACTTCTGCCTTCTCTGCAGTGA
3' ss Score
7.91
5' ss Seq
CAGGTGAGT
5' ss Score
10.67
Exon sequences
Seq C1 exon
GTGGCCGGACTGTGCCATGCTGCTCCGGACCCAAGGTGGCCGCTCTCACTGTGGGGACCGTCCTGCTCCTGACAGGCATCGGGGCAGCGTCCTGGGCCATTG
Seq A exon
TGACTGTTCTACTCAGGAGTGATCAGGAGCCACTGTATCCAG
Seq C2 exon
TGCAGGTCGGCCCGGCGGATGCTCGGCTCACGGTGTTCGACCAGACAGAGGGCACGTGGCGCCTGCTTTGCTCCTCGCGCTCCAATGCCAGGGTGGCGGGGCTCAGCTGCGAGGAGATGGGCTTCCTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000003829_MULTIEX1-2/2=1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
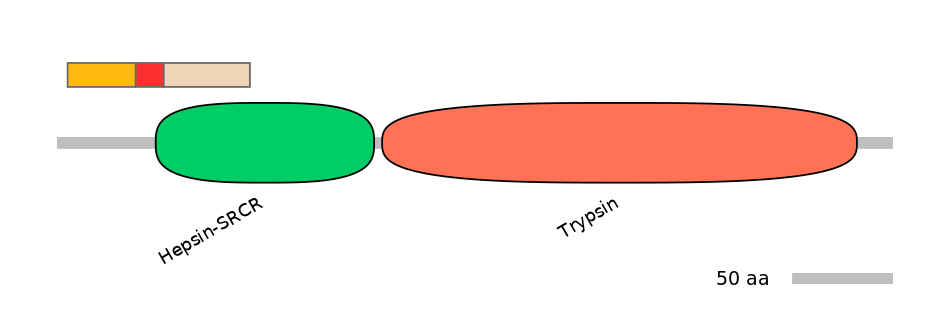
Domain overlap (PFAM):
C1:
NO
A:
PF092725=Hepsin-SRCR=PU(3.6=26.7)
C2:
PF092725=Hepsin-SRCR=FE(39.1=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CAGCGTCCTGGGCCATTG
R:
CTGAGGAAGCCCATCTCCTCG
Band lengths:
148-190
Functional annotations
There are 1 annotated functions for this event
PMID: 15627507
Both exons together encode a transmembrane domain and cause cytosolic retention when skipped. Hepsin/-TM (skipping) overexpression does not affect the growth of a human cancer cell line.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development