HsaEX0030402 @ hg38
Exon Skipping
Gene
ENSG00000105707 | HPN
Description
hepsin [Source:HGNC Symbol;Acc:HGNC:5155]
Coordinates
chr19:35049290-35059802:+
Coord C1 exon
chr19:35049290-35049391
Coord A exon
chr19:35049475-35049516
Coord C2 exon
chr19:35059673-35059802
Length
42 bp
Sequences
Splice sites
3' ss Seq
CACCCCTCCCTTCTCTGCAGTGG
3' ss Score
9.17
5' ss Seq
CAGGTGAGT
5' ss Score
10.67
Exon sequences
Seq C1 exon
GTGGCCGGACTGTGCCATGCTGCTCCAGACCCAAGGTGGCAGCTCTCACTGCGGGGACCCTGCTACTTCTGACAGCCATCGGGGCGGCATCCTGGGCCATTG
Seq A exon
TGGCTGTTCTCCTCAGGAGTGACCAGGAGCCGCTGTACCCAG
Seq C2 exon
TGCAGGTCAGCTCTGCGGACGCTCGGCTCATGGTCTTTGACAAGACGGAAGGGACGTGGCGGCTGCTGTGCTCCTCGCGCTCCAACGCCAGGGTAGCCGGACTCAGCTGCGAGGAGATGGGCTTCCTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000105707_MULTIEX1-3/4=2-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.036 A=0.067 C2=0.000
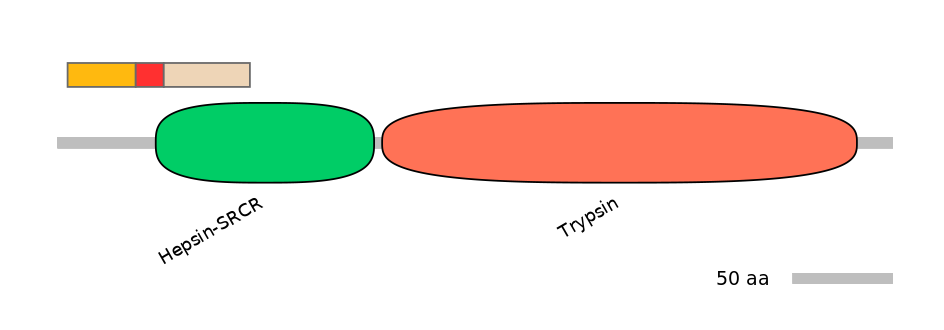
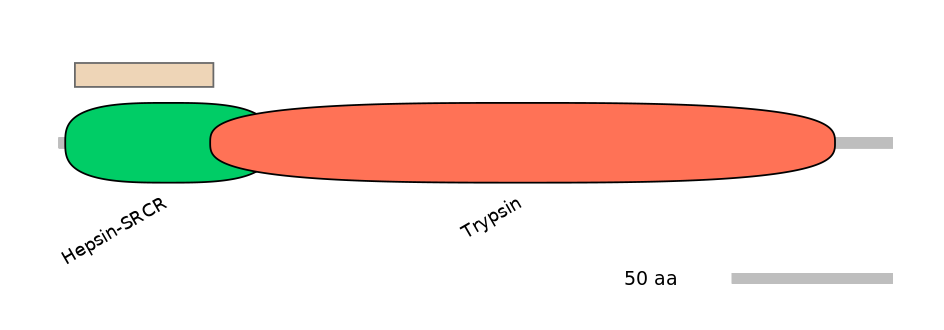
Domain overlap (PFAM):
C1:
NO
A:
PF092725=Hepsin-SRCR=PU(12.9=26.7)
C2:
PF092725=Hepsin-SRCR=FE(66.2=100),PF0008921=Trypsin=PU(0.5=2.3)
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CATGCTGCTCCAGACCCAAG
R:
CGTCCCTTCCGTCTTGTCAAA
Band lengths:
143-185
Functional annotations
There are 1 annotated functions for this event
PMID: 15627507
Both exons together encode a transmembrane domain and cause cytosolic retention when skipped. Hepsin/-TM (skipping) overexpression does not affect the growth of a human cancer cell line.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development