BtaEX6046313 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000044129 | ST6GALNAC6
Description
Bos taurus ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1, 3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 (ST6GALNAC6), mRNA. [Source:RefSeq mRNA;Acc:NM_001001151]
Coordinates
chr11:98575105-98579039:-
Coord C1 exon
chr11:98579014-98579039
Coord A exon
chr11:98577315-98577402
Coord C2 exon
chr11:98575105-98575284
Length
88 bp
Sequences
Splice sites
3' ss Seq
ATCCTTTCTGTGGCTTGCAGGTG
3' ss Score
10.1
5' ss Seq
AAAGTAAGT
5' ss Score
9.72
Exon sequences
Seq C1 exon
ATGGCTTGCCCGAGGCCCCTCAGCCA
Seq A exon
GTGTGACCACACACCCCTACCTGGGCCACCTGCAGGACACTGGCCCCTGCCCCTCAGCAGACGCCGGAGAGAGATGAAGAGCAACAAA
Seq C2 exon
GAGCAGCGGTCAGCAGTGTTCGTGATCCTCTTTGCCCTCATCACCATCCTCATCCTCTACAGCTCCAGCAGTGCCAATGAGGTCTTTCATTATGGCTCCCTGCGGGGCCGGACACGCCGGCCTGTCAACCTCAGGAAGTGGAGCATCACTGACGGATACATCCCCATTCTTGGCAACAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000044129-'3-6,'3-3,9-6=AN
Average complexity
A_C2
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.333 A=0.722 C2=0.000
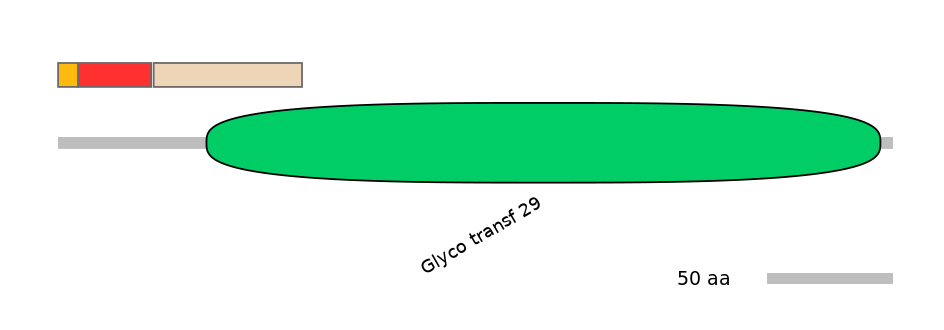
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0077713=Glyco_transf_29=PU(14.1=63.3)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]