DreEX6030711 @ danRer10
Exon Skipping
Gene
ENSDARG00000009026 | CR388002.1
Description
Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F4G7]
Coordinates
chr1:13423894-13425115:+
Coord C1 exon
chr1:13423894-13423992
Coord A exon
chr1:13424837-13424935
Coord C2 exon
chr1:13425017-13425115
Length
99 bp
Sequences
Splice sites
3' ss Seq
TTTTTTCTTTGTACATTCAGAAT
3' ss Score
8.46
5' ss Seq
ATGGTATGA
5' ss Score
6.56
Exon sequences
Seq C1 exon
TTGGGCTACACTCCTCTTATCGTGGCTTGCCACTATGGAAATGTCAAAATGGTTAATTTCCTTTTGCAAAATGGCGCTAATGTTAATGGAAAAACAAAG
Seq A exon
AATGGTTACACACCACTTCATCAAGCTGCTCAACAGGGAAACACACACATTGTCAATGTCCTCCTGCAGCACGGAGCTAAGCCTAATGCTGTTACCATG
Seq C2 exon
AACGGAAACACTGCACTCTCTATTGCGAAGCGTCTGGGGTACATCTCAGTGGTGGACACTCTGAAAGTTGTTACTGAGGAAATCATCACAACCACAACT
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000009026-'23-24,'23-23,24-24=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.091 A=0.303 C2=0.030
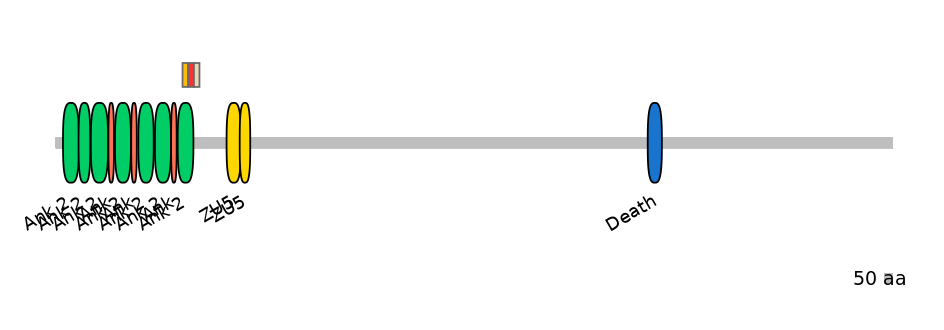
Domain overlap (PFAM):
C1:
PF127962=Ank_2=FE(34.8=100)
A:
PF127962=Ank_2=PD(32.6=90.9)
C2:
NO
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGGCTACACTCCTCTTATCGT
R:
ACAACTTTCAGAGTGTCCACCAC
Band lengths:
169-268
Functional annotations
There are 1 annotated functions for this event
PMID: 14722080
Exon 21 (HsaEX0004130) encodes ankyrin repeat 23. InsP3R-Ankyrin-B Interaction Requires Tips of beta-Hairpin Loops of ANK Repeats 22-24, and therefore it is expected to be affected by skipping. The function of Hsa0004130 has not been examined in isolation.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]