GgaEX1065928 @ galGal4
Exon Skipping
Gene
ENSGALG00000012044 | chAnk2
Description
Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1NG08]
Coordinates
chr4:56410837-56416722:-
Coord C1 exon
chr4:56416624-56416722
Coord A exon
chr4:56413902-56414000
Coord C2 exon
chr4:56410837-56410935
Length
99 bp
Sequences
Splice sites
3' ss Seq
GTAACCACCATGGATTACAGAAT
3' ss Score
1.12
5' ss Seq
ACCGTAAGT
5' ss Score
10.43
Exon sequences
Seq C1 exon
CTTGGTTATACACCTTTAATTGTGGCCTGTCACTATGGAAACATCAAAATGGTGAATTTTCTTCTCAAGCAGGGAGCAAATGTCAATGCTAAAACAAAG
Seq A exon
AATGGGTACACTCCTCTCCACCAAGCTGCTCAACAAGGTCACACTCACATCATTAATGTCCTTCTCCAACATGGAGCTAAGCCCAATGCCATCACCACC
Seq C2 exon
AATGGTAACACTGCCTTAGCTATTGCTAGGCGTCTGGGATATATCTCAGTGGTCGATACGCTGAAGGTTGTAACGGAGGAGATTACTACCACCACAACT
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000012044-'35-36,'35-35,36-36
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.061 A=0.303 C2=0.124
Domain overlap (PFAM):
C1:
PF127962=Ank_2=PD(44.9=93.9),PF127962=Ank_2=PU(3.1=6.1)
A:
PF127962=Ank_2=PD(31.9=87.9)
C2:
PF127962=Ank_2=PD(42.2=81.8)
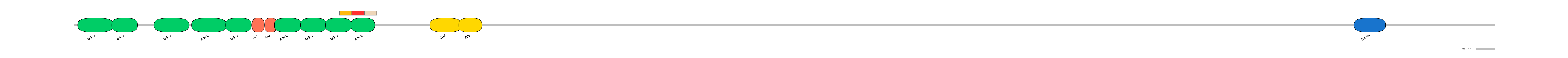
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACACCTTTAATTGTGGCCTGTCA
R:
CCTCCGTTACAACCTTCAGCG
Band lengths:
169-268
Functional annotations
There are 1 annotated functions for this event
PMID: 14722080
Exon 21 (HsaEX0004130) encodes ankyrin repeat 23. InsP3R-Ankyrin-B Interaction Requires Tips of beta-Hairpin Loops of ANK Repeats 22-24, and therefore it is expected to be affected by skipping. The function of Hsa0004130 has not been examined in isolation.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]